Note
Go to the end to download the full example code.
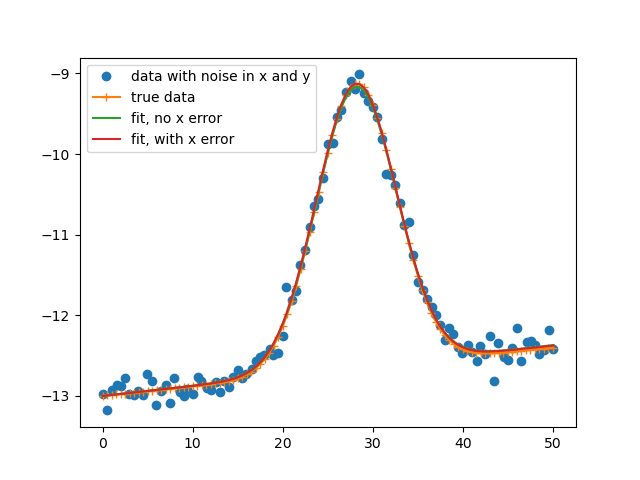
Fitting data with uncertainties in x and y¶
This examples shows a general way of fitting a model to y(x) data which has uncertainties in both y and x.
- For more in-depth discussion, see
### not including uncertainty in x:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 62
# data points = 101
# variables = 5
chi-square = 344.228513
reduced chi-square = 3.58571368
Akaike info crit = 133.844705
Bayesian info crit = 146.920308
[[Variables]]
amp: 38.7909094 +/- 0.54820333 (1.41%) (init = 50)
cen: 28.1858429 +/- 0.05334483 (0.19%) (init = 25)
sig: 4.44210577 +/- 0.05938737 (1.34%) (init = 10)
slope: 0.01260816 +/- 8.1762e-04 (6.48%) (init = 0.0001)
offset: -13.0004948 +/- 0.02386032 (0.18%) (init = -5)
[[Correlations]] (unreported correlations are < 0.100)
C(slope, offset) = -0.7603
C(amp, sig) = +0.7056
C(amp, offset) = -0.2735
C(cen, slope) = -0.2460
C(amp, slope) = -0.2115
C(sig, offset) = -0.1924
C(cen, offset) = +0.1870
C(sig, slope) = -0.1495
None
### including uncertainty in x:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 43
# data points = 101
# variables = 5
chi-square = 223.406586
reduced chi-square = 2.32715193
Akaike info crit = 90.1811577
Bayesian info crit = 103.256760
[[Variables]]
amp: 39.2776438 +/- 0.94011531 (2.39%) (init = 50)
cen: 28.1731192 +/- 0.12633327 (0.45%) (init = 25)
sig: 4.44581423 +/- 0.09870523 (2.22%) (init = 10)
slope: 0.01252275 +/- 6.7955e-04 (5.43%) (init = 0.0001)
offset: -13.0005237 +/- 0.01980939 (0.15%) (init = -5)
[[Correlations]] (unreported correlations are < 0.100)
C(amp, sig) = +0.8188
C(slope, offset) = -0.7475
C(amp, offset) = -0.2037
C(cen, slope) = -0.1715
C(sig, offset) = -0.1655
C(amp, slope) = -0.1630
C(cen, offset) = +0.1381
C(sig, slope) = -0.1359
None
[0.01438036 0.52896805 1.04666251 1.58348091]
import matplotlib.pyplot as plt
import numpy as np
from lmfit import Minimizer, Parameters, report_fit
from lmfit.lineshapes import gaussian
# create data to be fitted
np.random.seed(17)
xtrue = np.linspace(0, 50, 101)
xstep = xtrue[1] - xtrue[0]
amp, cen, sig, offset, slope = 39, 28.2, 4.4, -13, 0.012
ytrue = (gaussian(xtrue, amplitude=amp, center=cen, sigma=sig)
+ offset + slope * xtrue)
ydat = ytrue + np.random.normal(size=xtrue.size, scale=0.1)
# we add errors to x after y has been created, as if there is
# an ideal y(x) and we have noise in both x and y.
# we force the uncertainty away from 'normal', forcing
# it to be smaller than the step size.
xerr = np.random.normal(size=xtrue.size, scale=0.1*xstep)
max_xerr = 0.8*xstep
xerr[np.where(xerr > max_xerr)] = max_xerr
xerr[np.where(xerr < -max_xerr)] = -max_xerr
xdat = xtrue + xerr
# now we assert that we know the uncertaintits in y and x
# we'll pick values that are reesonable but not exactly
# what we used to make the noise
yerr = 0.06
xerr = xstep
def peak_model(params, x):
"""Model a peak with a linear background."""
amp = params['amp'].value
cen = params['cen'].value
sig = params['sig'].value
offset = params['offset'].value
slope = params['slope'].value
return offset + slope * x + gaussian(x, amplitude=amp, center=cen, sigma=sig)
# objective without xerr
def objective_no_xerr(params, x, y, yerr):
model = peak_model(params, x)
return (model - y) / abs(yerr)
# objective with xerr
def objective_with_xerr(params, x, y, yerr, xerr):
model = peak_model(params, x)
dmodel_dx = np.gradient(model) / np.gradient(x)
dmodel = np.sqrt(yerr**2 + (xerr*dmodel_dx)**2)
return (model - y) / dmodel
# create a set of Parameters
params = Parameters()
params.add('amp', value=50, min=0)
params.add('cen', value=25)
params.add('sig', value=10)
params.add('slope', value=1.e-4)
params.add('offset', value=-5)
# first fit without xerr
mini1 = Minimizer(objective_no_xerr, params, fcn_args=(xdat, ydat, yerr))
result1 = mini1.minimize()
bestfit1 = peak_model(result1.params, xdat)
mini2 = Minimizer(objective_with_xerr, params, fcn_args=(xdat, ydat, yerr, xerr))
result2 = mini2.minimize()
bestfit2 = peak_model(result2.params, xdat)
print("### not including uncertainty in x:")
print(report_fit(result1))
print("### including uncertainty in x:")
print(report_fit(result2))
print(xdat[:4])
plt.plot(xdat, ydat, 'o', label='data with noise in x and y')
plt.plot(xtrue, ytrue, '-+', label='true data')
plt.plot(xdat, bestfit1, label='fit, no x error')
plt.plot(xdat, bestfit2, label='fit, with x error')
plt.legend()
plt.show()
# # <end examples/doc_uncertainties_in_x_and_y.py>
Total running time of the script: (0 minutes 0.311 seconds)
