Note
Go to the end to download the full example code.
Emcee and the Model Interface¶
import corner
import matplotlib.pyplot as plt
import numpy as np
import lmfit
Set up a double-exponential function and create a Model:
Generate some fake data from the model with added noise:
truths = (3.0, 2.0, -5.0, 10.0)
x = np.linspace(1, 10, 250)
np.random.seed(0)
y = double_exp(x, *truths)+0.1*np.random.randn(x.size)
Create model parameters and give them initial values:
p = model.make_params(a1=4, t1=3, a2=4, t2=3)
Fit the model using a traditional minimizer, and show the output:
[[Fit Statistics]]
# fitting method = Nelder-Mead
# function evals = 609
# data points = 250
# variables = 4
chi-square = 2.33333982
reduced chi-square = 0.00948512
Akaike info crit = -1160.54007
Bayesian info crit = -1146.45423
R-squared = 0.94237407
[[Variables]]
a1: 2.98623689 +/- 0.15010519 (5.03%) (init = 4)
t1: 1.30993186 +/- 0.13449656 (10.27%) (init = 3)
a2: -4.33525597 +/- 0.11765824 (2.71%) (init = 4)
t2: 11.8240752 +/- 0.47172610 (3.99%) (init = 3)
[[Correlations]] (unreported correlations are < 0.100)
C(a2, t2) = +0.9876
C(t1, a2) = -0.9278
C(t1, t2) = -0.8852
C(a1, t1) = -0.6093
C(a1, a2) = +0.2973
C(a1, t2) = +0.2319
Calculate parameter covariance using emcee:
start the walkers out at the best-fit values
set
is_weightedtoFalseto estimate the noise weightsset some sensible priors on the uncertainty to keep the MCMC in check
run the MCMC algorithm and show the results:
lmfit.report_fit(result_emcee)
[[Fit Statistics]]
# fitting method = emcee
# function evals = 500000
# data points = 250
# variables = 5
chi-square = 245.221790
reduced chi-square = 1.00090526
Akaike info crit = 5.17553688
Bayesian info crit = 22.7828415
R-squared = 0.94233862
[[Variables]]
a1: 2.99546858 +/- 0.14834594 (4.95%) (init = 2.986237)
t1: 1.32127391 +/- 0.14079400 (10.66%) (init = 1.309932)
a2: -4.34376940 +/- 0.12389335 (2.85%) (init = -4.335256)
t2: 11.7937607 +/- 0.48879633 (4.14%) (init = 11.82408)
__lnsigma: -2.32712371 +/- 0.04514314 (1.94%) (init = -2.302585)
[[Correlations]] (unreported correlations are < 0.100)
C(a2, t2) = +0.9810
C(t1, a2) = -0.9367
C(t1, t2) = -0.8949
C(a1, t1) = -0.5154
C(a1, a2) = +0.2197
C(a1, t2) = +0.1868
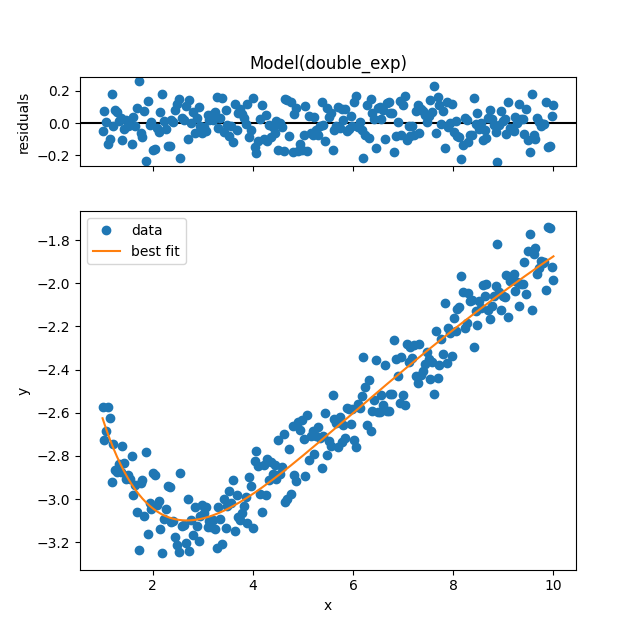
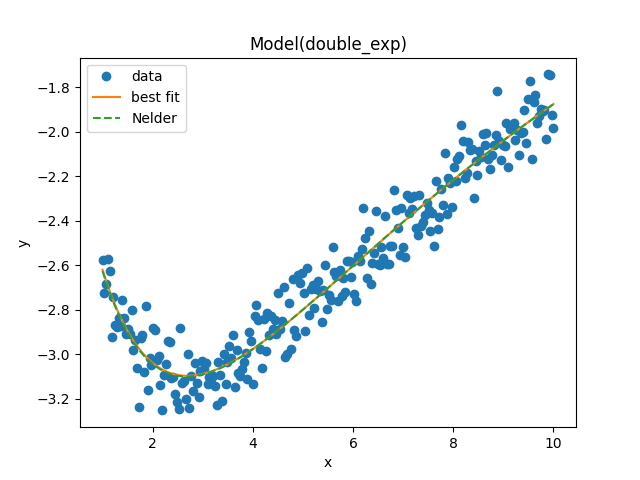
result_emcee.plot_fit()
plt.plot(x, model.eval(params=result.params, x=x), '--', label='Nelder')
plt.legend()
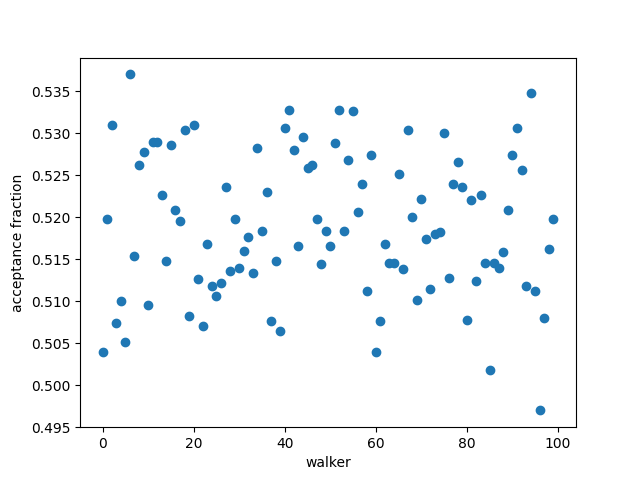
Check the acceptance fraction to see whether emcee performed well:
plt.plot(result_emcee.acceptance_fraction, 'o')
plt.xlabel('walker')
plt.ylabel('acceptance fraction')
Try to compute the autocorrelation time:
if hasattr(result_emcee, "acor"):
print("Autocorrelation time for the parameters:")
print("----------------------------------------")
for i, p in enumerate(result.params):
print(f'{p} = {result_emcee.acor[i]:.3f}')
Autocorrelation time for the parameters:
----------------------------------------
a1 = 61.334
t1 = 85.867
a2 = 86.046
t2 = 84.745
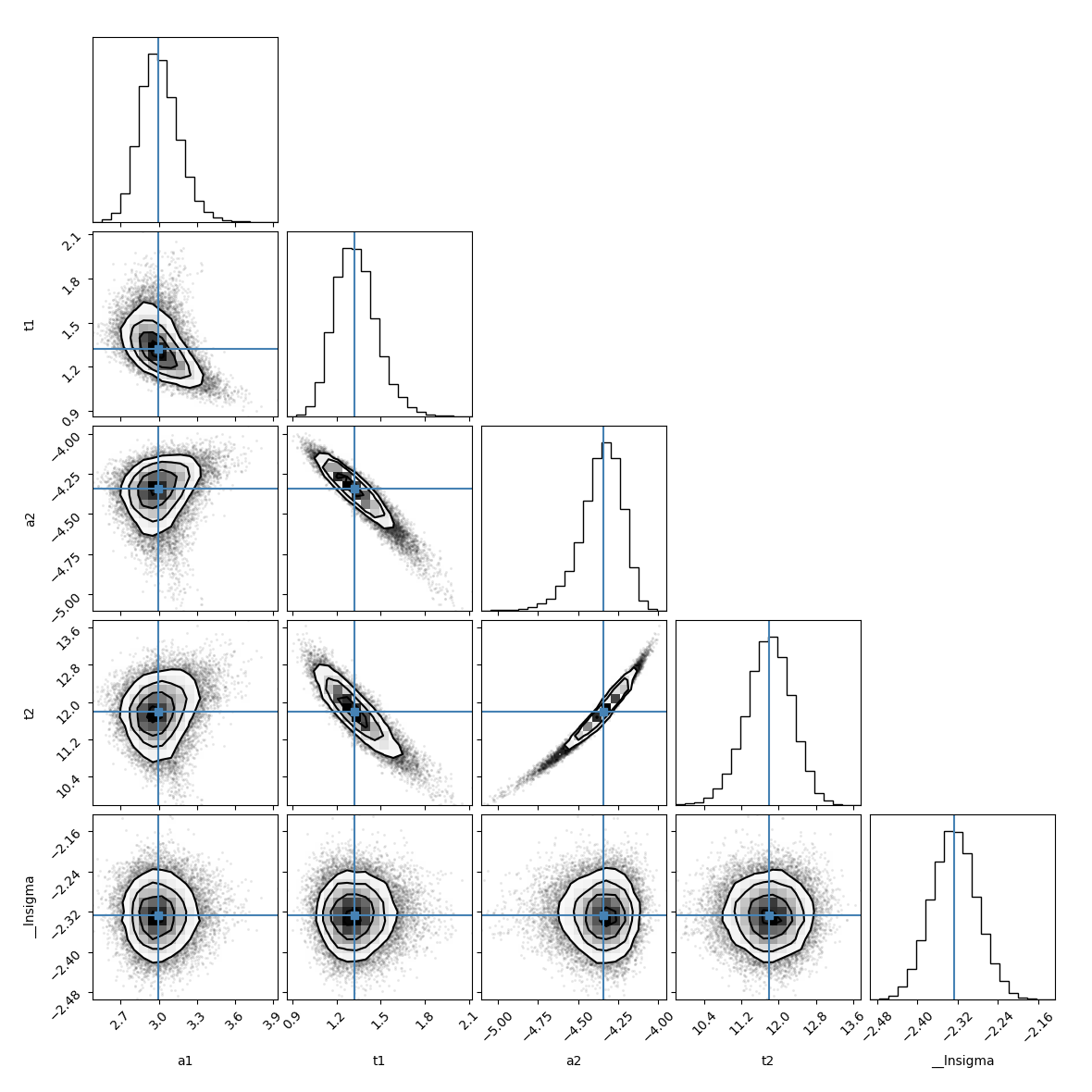
Plot the parameter covariances returned by emcee using corner:
emcee_corner = corner.corner(result_emcee.flatchain, labels=result_emcee.var_names,
truths=list(result_emcee.params.valuesdict().values()))
print("\nmedian of posterior probability distribution")
print('--------------------------------------------')
lmfit.report_fit(result_emcee.params)
median of posterior probability distribution
--------------------------------------------
[[Variables]]
a1: 2.99546858 +/- 0.14834594 (4.95%) (init = 2.986237)
t1: 1.32127391 +/- 0.14079400 (10.66%) (init = 1.309932)
a2: -4.34376940 +/- 0.12389335 (2.85%) (init = -4.335256)
t2: 11.7937607 +/- 0.48879633 (4.14%) (init = 11.82408)
__lnsigma: -2.32712371 +/- 0.04514314 (1.94%) (init = -2.302585)
[[Correlations]] (unreported correlations are < 0.100)
C(a2, t2) = +0.9810
C(t1, a2) = -0.9367
C(t1, t2) = -0.8949
C(a1, t1) = -0.5154
C(a1, a2) = +0.2197
C(a1, t2) = +0.1868
Find the maximum likelihood solution:
highest_prob = np.argmax(result_emcee.lnprob)
hp_loc = np.unravel_index(highest_prob, result_emcee.lnprob.shape)
mle_soln = result_emcee.chain[hp_loc]
print("\nMaximum Likelihood Estimation (MLE):")
print('----------------------------------')
for ix, param in enumerate(emcee_params):
print(f"{param}: {mle_soln[ix]:.3f}")
quantiles = np.percentile(result_emcee.flatchain['t1'], [2.28, 15.9, 50, 84.2, 97.7])
print(f"\n\n1 sigma spread = {0.5 * (quantiles[3] - quantiles[1]):.3f}")
print(f"2 sigma spread = {0.5 * (quantiles[4] - quantiles[0]):.3f}")
Maximum Likelihood Estimation (MLE):
----------------------------------
a1: 2.971
t1: 1.317
a2: -4.336
t2: 11.815
__lnsigma: -2.336
1 sigma spread = 0.141
2 sigma spread = 0.291
Total running time of the script: (0 minutes 45.014 seconds)
