Calculation of confidence intervals¶
The lmfit confidence module allows you to explicitly calculate
confidence intervals for variable parameters. For most models, it is not
necessary since the estimation of the standard error from the estimated
covariance matrix is normally quite good.
But for some models, the sum of two exponentials for example, the approximation
begins to fail. For this case, lmfit has the function conf_interval()
to calculate confidence intervals directly. This is substantially slower
than using the errors estimated from the covariance matrix, but the results
are more robust.
Please note that conf_interval() is not suited to work with
fit results obtained by emcee.
Method used for calculating confidence intervals¶
The F-test is used to compare our null model, which is the best fit we have found, with an alternate model, where one of the parameters is fixed to a specific value. The value is changed until the difference between \(\chi^2_0\) and \(\chi^2_{f}\) can’t be explained by the loss of a degree of freedom within a certain confidence.
N is the number of data points and P the number of parameters of the null model.
\(P_{fix}\) is the number of fixed parameters (or to be more clear, the
difference of number of parameters between our null model and the alternate
model).
Adding a log-likelihood method is under consideration.
A basic example¶
First we create an example problem:
import numpy as np
import lmfit
x = np.linspace(0.3, 10, 100)
np.random.seed(0)
y = 1/(0.1*x) + 2 + 0.1*np.random.randn(x.size)
pars = lmfit.Parameters()
pars.add_many(('a', 0.1), ('b', 1))
def residual(p):
return 1/(p['a']*x) + p['b'] - y
before we can generate the confidence intervals, we have to run a fit, so that the automated estimate of the standard errors can be used as a starting point:
mini = lmfit.Minimizer(residual, pars)
result = mini.minimize()
print(lmfit.fit_report(result.params))
[[Variables]]
a: 0.09943896 +/- 1.9322e-04 (0.19%) (init = 0.1)
b: 1.98476942 +/- 0.01222678 (0.62%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(a, b) = +0.6008
Now it is just a simple function call to calculate the confidence intervals:
ci = lmfit.conf_interval(mini, result)
lmfit.printfuncs.report_ci(ci)
99.73% 95.45% 68.27% _BEST_ 68.27% 95.45% 99.73%
a: -0.00059 -0.00039 -0.00019 0.09944 +0.00019 +0.00039 +0.00060
b: -0.03764 -0.02477 -0.01229 1.98477 +0.01229 +0.02477 +0.03764
This shows the best-fit values for the parameters in the _BEST_ column,
and parameter values that are at the varying confidence levels given by
steps in \(\sigma\). As we can see, the estimated error is almost the
same, and the uncertainties are well behaved: Going from 1-\(\sigma\)
(68% confidence) to 3-\(\sigma\) (99.7% confidence) uncertainties is
fairly linear. It can also be seen that the errors are fairly symmetric
around the best fit value. For this problem, it is not necessary to
calculate confidence intervals, and the estimates of the uncertainties from
the covariance matrix are sufficient.
Working without standard error estimates¶
Sometimes the estimation of the standard errors from the covariance matrix fails, especially if values are near given bounds. Hence, to find the confidence intervals in these cases, it is necessary to set the errors by hand. Note that the standard error is only used to find an upper limit for each value, hence the exact value is not important.
To set the step-size to 10% of the initial value we loop through all parameters and set it manually:
for p in result.params:
result.params[p].stderr = abs(result.params[p].value * 0.1)
Calculating and visualizing maps of \(\chi^2\)¶
The estimated values for the \(1-\sigma\) standard error calculated by default for each fit include the effects of correlation between pairs of variables, but assumes the uncertainties are symmetric. While it doesn’t exactly say what the values of the \(n-\sigma\) uncertainties would be, the implication is that the \(n-\sigma\) error is simply \(n^2\sigma\).
The conf_interval() function described above improves on these
automatically (and quickly) calculated uncertainies by explicitly finding
\(n-\sigma\) confidence levels in both directions – it does not assume
that the uncertainties are symmetric. This function also takes into account the
correlations between pairs of variables, but it does not convey this
information very well.
For even further exploration of the confidence levels of parameter values, it can be useful to calculate maps of \(\chi^2\) values for pairs of variables around their best fit values and visualize these as contour plots. Typically, pairs of variables will have elliptical contours of constant \(n-\sigma\) level, with highly-correlated pairs of variables having high ratios of major and minor axes.
The conf_interval2d() can calculate 2-d arrays or maps of either
probability or \(\delta \chi^2 = \chi^2 - \chi_{\mathrm{best}}^2\) for any
pair of variables. Visualizing these can help better understand the nature of
the uncertainties and correlations between parameters. To illustrate this,
we’ll start with an example fit to data that we deliberately add components not
accounted for in the model, and with slightly non-Gaussian noise – a
constructed but “real-world” example:
# <examples/doc_confidence_chi2_maps.py>
import matplotlib.pyplot as plt
import numpy as np
from lmfit import conf_interval, conf_interval2d, report_ci
from lmfit.lineshapes import gaussian
from lmfit.models import GaussianModel, LinearModel
sigma_levels = [1, 2, 3]
rng = np.random.default_rng(seed=102)
#########################
# set up data -- deliberately adding imperfections and
# a small amount of non-Gaussian noise
npts = 501
x = np.linspace(1, 100, num=npts)
noise = rng.normal(scale=0.3, size=npts) + 0.2*rng.f(3, 9, size=npts)
y = (gaussian(x, amplitude=83, center=47., sigma=5.)
+ 0.02*x + 4 + 0.25*np.cos((x-20)/8.0) + noise)
mod = GaussianModel() + LinearModel()
params = mod.make_params(amplitude=100, center=50, sigma=5,
slope=0, intecept=2)
out = mod.fit(y, params, x=x)
print(out.fit_report())
#########################
# run conf_intervale, print report
sigma_levels = [1, 2, 3]
ci = conf_interval(out, out, sigmas=sigma_levels)
print("## Confidence Report:")
report_ci(ci)
[[Model]]
(Model(gaussian) + Model(linear))
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 31
# data points = 501
# variables = 5
chi-square = 103.861381
reduced chi-square = 0.20939794
Akaike info crit = -778.348033
Bayesian info crit = -757.265003
R-squared = 0.93782756
[[Variables]]
amplitude: 78.8171374 +/- 1.21910939 (1.55%) (init = 100)
center: 47.0751649 +/- 0.07576660 (0.16%) (init = 50)
sigma: 4.93298753 +/- 0.07984021 (1.62%) (init = 5)
slope: 0.01839006 +/- 7.1957e-04 (3.91%) (init = 0)
intercept: 4.39234411 +/- 0.04420227 (1.01%) (init = 0)
fwhm: 11.6162977 +/- 0.18800933 (1.62%) == '2.3548200*sigma'
height: 6.37412722 +/- 0.08603873 (1.35%) == '0.3989423*amplitude/max(1e-15, sigma)'
[[Correlations]] (unreported correlations are < 0.100)
C(slope, intercept) = -0.8421
C(amplitude, sigma) = +0.6371
C(amplitude, intercept) = -0.3373
C(sigma, intercept) = -0.2149
C(center, slope) = -0.1026
## Confidence Report:
99.73% 95.45% 68.27% _BEST_ 68.27% 95.45% 99.73%
amplitude: -3.62610 -2.41983 -1.21237 78.81714 +1.22111 +2.45479 +3.70515
center : -0.22849 -0.15214 -0.07584 47.07516 +0.07587 +0.15225 +0.22873
sigma : -0.23335 -0.15640 -0.07870 4.93299 +0.08000 +0.16158 +0.24509
slope : -0.00217 -0.00144 -0.00072 0.01839 +0.00072 +0.00144 +0.00217
intercept: -0.13326 -0.08860 -0.04423 4.39234 +0.04421 +0.08854 +0.13312
The reports show that we obtained a pretty good fit, and that the automated estimates of the uncertainties are actually pretty good – agreeing to the second decimal place. But we also see that some of the uncertainties do become noticeably asymmetric at high \(n-\sigma\) levels.
We’ll plot this data and fit, and then further explore these uncertainties
using conf_interval2d(). Please note that conf_interval2d() shows the
confidence intervals obtained considering the standard errors, not those obtained
by ‘conf_interval(out, out, sigmas=sigma_levels)’.
#########################
# plot initial fit
colors = ('#2030b0', '#b02030', '#207070')
fig, axes = plt.subplots(2, 3, figsize=(15, 9.5))
axes[0, 0].plot(x, y, 'o', markersize=3, label='data', color=colors[0])
axes[0, 0].plot(x, out.best_fit, label='fit', color=colors[1])
axes[0, 0].set_xlabel('x')
axes[0, 0].set_ylabel('y')
axes[0, 0].legend()
aix, aiy = 0, 0
nsamples = 50
explicitly_calculate_sigma = True
for pairs in (('sigma', 'amplitude'), ('intercept', 'amplitude'),
('slope', 'intercept'), ('slope', 'center'), ('sigma', 'center')):
xpar, ypar = pairs
if explicitly_calculate_sigma:
print(f"Generating chi-square map for {pairs}")
c_x, c_y, chi2_mat = conf_interval2d(out, out, xpar, ypar,
nsamples, nsamples, nsigma=3.5,
chi2_out=True)
# explicitly calculate sigma matrix: sigma increases chi_square
# from chi_square_best
# to chi_square + sigma**2 * reduced_chi_square
# so: sigma = sqrt((chi2-chi2_best)/ reduced_chi_square)
chi2_min = chi2_mat.min()
sigma_mat = np.sqrt((chi2_mat-chi2_min)/out.redchi)
else:
print(f"Generating sigma map for {pairs}")
# or, you could just calculate the matrix of probabilities as:
c_x, c_y, sigma_mat = conf_interval2d(out, out, xpar, ypar,
nsamples, nsamples, nsigma=3.5)
aix += 1
if aix == 2:
aix = 0
aiy += 1
ax = axes[aix, aiy]
cnt = ax.contour(c_x, c_y, sigma_mat, levels=sigma_levels, colors=colors,
linestyles='-')
ax.clabel(cnt, inline=True, fmt=r"$\sigma=%.0f$", fontsize=13)
# draw boxes for estimated uncertaties:
# dotted : scaled stderr from initial fit
# dashed : values found from conf_interval()
xv = out.params[xpar].value
xs = out.params[xpar].stderr
yv = out.params[ypar].value
ys = out.params[ypar].stderr
cix = ci[xpar]
ciy = ci[ypar]
nc = len(sigma_levels)
for i in sigma_levels:
# dotted line: scaled stderr
ax.plot((xv-i*xs, xv+i*xs, xv+i*xs, xv-i*xs, xv-i*xs),
(yv-i*ys, yv-i*ys, yv+i*ys, yv+i*ys, yv-i*ys),
linestyle='dotted', color=colors[i-1])
# dashed line: refined uncertainties from conf_interval
xsp, xsm = cix[nc+i][1], cix[nc-i][1]
ysp, ysm = ciy[nc+i][1], ciy[nc-i][1]
ax.plot((xsm, xsp, xsp, xsm, xsm), (ysm, ysm, ysp, ysp, ysm),
linestyle='dashed', color=colors[i-1])
ax.set_xlabel(xpar)
ax.set_ylabel(ypar)
ax.grid(True, color='#d0d0d0')
plt.show()
# <end examples/doc_confidence_chi2_maps.py>
Generating chi-square map for ('sigma', 'amplitude')
Generating chi-square map for ('intercept', 'amplitude')
Generating chi-square map for ('slope', 'intercept')
Generating chi-square map for ('slope', 'center')
Generating chi-square map for ('sigma', 'center')
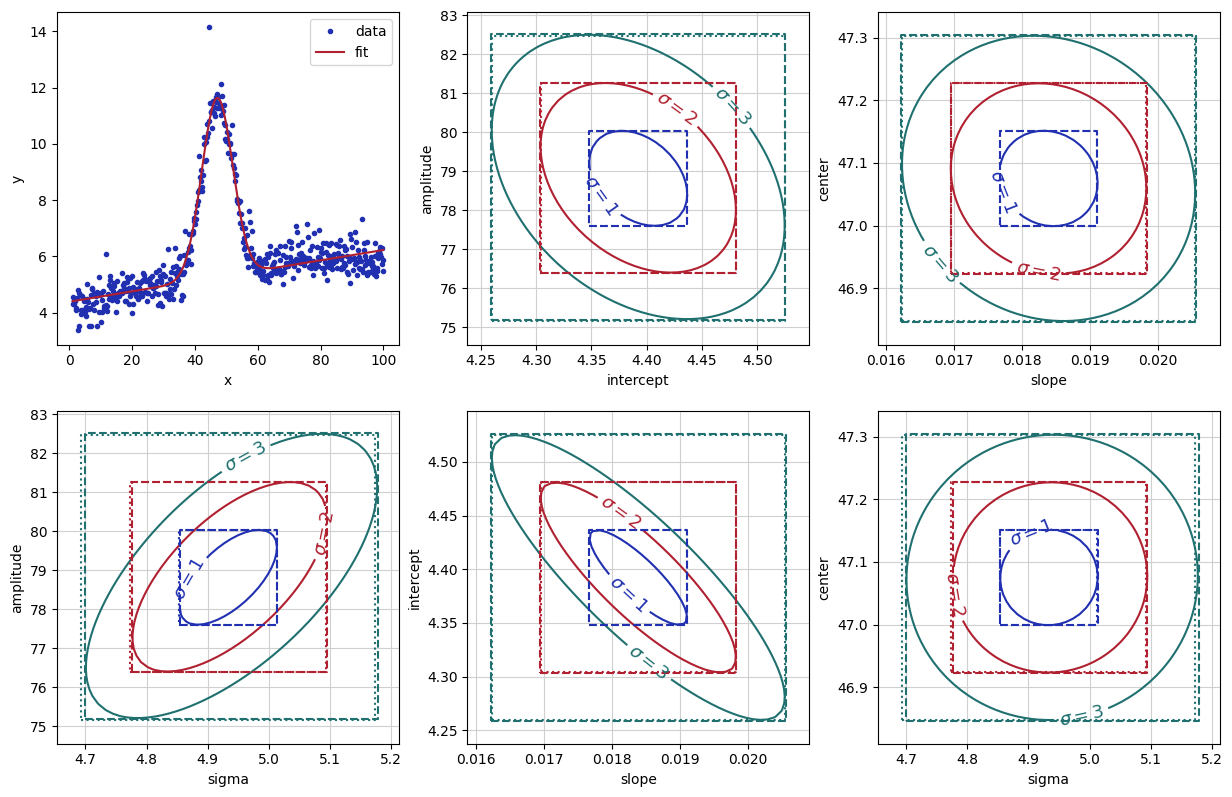
Here we made contours for the \(n-\sigma\) levels from the 2-D array of \(\chi^2\) by noting that the \(n-\sigma\) level will have \(\chi^2\) increased by \(n^2\chi_\nu^2\) where \(\chi_\nu^2\) is reduced chi-square.
The dotted boxes show both the scaled values of the standard errors from the
initial fit, and the dashed boxes show the confidence levels from
conf_interval(). You can see that the notion of increasing
\(\chi^2\) by \(\chi_\nu^2\) works very well, and that there is a small
asymmetry in the uncertainties for the amplitude and sigma parameters.
An advanced example for evaluating confidence intervals¶
Now we look at a problem where calculating the error from approximated
covariance can lead to misleading result – the same double exponential
problem shown in Minimizer.emcee() - calculating the posterior probability distribution of parameters. In fact such a problem is particularly
hard for the Levenberg-Marquardt method, so we first estimate the results
using the slower but robust Nelder-Mead method. We can then compare the
uncertainties computed (if the numdifftools package is installed) with
those estimated using Levenberg-Marquardt around the previously found
solution. We can also compare to the results of using emcee.
# <examples/doc_confidence_advanced.py>
import matplotlib.pyplot as plt
import numpy as np
import lmfit
x = np.linspace(1, 10, 250)
np.random.seed(0)
y = 3.0*np.exp(-x/2) - 5.0*np.exp(-(x-0.1)/10.) + 0.1*np.random.randn(x.size)
p = lmfit.create_params(a1=4, a2=4, t1=3, t2=3)
def residual(p):
return p['a1']*np.exp(-x/p['t1']) + p['a2']*np.exp(-(x-0.1)/p['t2']) - y
# create Minimizer
mini = lmfit.Minimizer(residual, p, nan_policy='propagate')
# first solve with Nelder-Mead algorithm
out1 = mini.minimize(method='Nelder')
# then solve with Levenberg-Marquardt using the
# Nelder-Mead solution as a starting point
out2 = mini.minimize(method='leastsq', params=out1.params)
lmfit.report_fit(out2.params, min_correl=0.5)
ci, trace = lmfit.conf_interval(mini, out2, sigmas=[1, 2], trace=True)
lmfit.printfuncs.report_ci(ci)
# plot data and best fit
plt.figure()
plt.plot(x, y)
plt.plot(x, residual(out2.params) + y, '-')
plt.show()
# plot confidence intervals (a1 vs t2 and a2 vs t2)
fig, axes = plt.subplots(1, 2, figsize=(12.8, 4.8))
cx, cy, grid = lmfit.conf_interval2d(mini, out2, 'a1', 't2', 30, 30)
ctp = axes[0].contourf(cx, cy, grid, np.linspace(0, 1, 11))
fig.colorbar(ctp, ax=axes[0])
axes[0].set_xlabel('a1')
axes[0].set_ylabel('t2')
cx, cy, grid = lmfit.conf_interval2d(mini, out2, 'a2', 't2', 30, 30)
ctp = axes[1].contourf(cx, cy, grid, np.linspace(0, 1, 11))
fig.colorbar(ctp, ax=axes[1])
axes[1].set_xlabel('a2')
axes[1].set_ylabel('t2')
plt.show()
# plot dependence between two parameters
fig, axes = plt.subplots(1, 2, figsize=(12.8, 4.8))
cx1, cy1, prob = trace['a1']['a1'], trace['a1']['t2'], trace['a1']['prob']
cx2, cy2, prob2 = trace['t2']['t2'], trace['t2']['a1'], trace['t2']['prob']
axes[0].scatter(cx1, cy1, c=prob, s=30)
axes[0].set_xlabel('a1')
axes[0].set_ylabel('t2')
axes[1].scatter(cx2, cy2, c=prob2, s=30)
axes[1].set_xlabel('t2')
axes[1].set_ylabel('a1')
plt.show()
# <end examples/doc_confidence_advanced.py>
which will report:
[[Variables]]
a1: 2.98622095 +/- 0.14867027 (4.98%) (init = 2.986237)
a2: -4.33526363 +/- 0.11527574 (2.66%) (init = -4.335256)
t1: 1.30994276 +/- 0.13121215 (10.02%) (init = 1.309932)
t2: 11.8240337 +/- 0.46316956 (3.92%) (init = 11.82408)
[[Correlations]] (unreported correlations are < 0.500)
C(a2, t2) = +0.9871
C(a2, t1) = -0.9246
C(t1, t2) = -0.8805
C(a1, t1) = -0.5988
95.45% 68.27% _BEST_ 68.27% 95.45%
a1: -0.27285 -0.14165 2.98622 +0.16354 +0.36343
a2: -0.30440 -0.13219 -4.33526 +0.10689 +0.19684
t1: -0.23392 -0.12494 1.30994 +0.14660 +0.32369
t2: -1.01937 -0.48813 11.82403 +0.46045 +0.90439
Again we called conf_interval(), this time with tracing and only for
1- and 2-\(\sigma\). Comparing these two different estimates, we see
that the estimate for a1 is reasonably well approximated from the
covariance matrix, but the estimates for a2 and especially for t1, and
t2 are very asymmetric and that going from 1 \(\sigma\) (68%
confidence) to 2 \(\sigma\) (95% confidence) is not very predictable.
Plots of the confidence region are shown in the figures below for a1 and
t2 (left), and a2 and t2 (right):
Neither of these plots is very much like an ellipse, which is implicitly
assumed by the approach using the covariance matrix. The plots actually
look quite a bit like those found with MCMC and shown in the “corner plot”
in Minimizer.emcee() - calculating the posterior probability distribution of parameters. In fact, comparing the confidence interval results
here with the results for the 1- and 2-\(\sigma\) error estimated with
emcee, we can see that the agreement is pretty good and that the
asymmetry in the parameter distributions are reflected well in the
asymmetry of the uncertainties.
The trace returned as the optional second argument from
conf_interval() contains a dictionary for each variable parameter.
The values are dictionaries with arrays of values for each variable, and an
array of corresponding probabilities for the corresponding cumulative
variables. This can be used to show the dependence between two
parameters:
fig, axes = plt.subplots(1, 2, figsize=(12.8, 4.8))
cx1, cy1, prob = trace['a1']['a1'], trace['a1']['t2'], trace['a1']['prob']
cx2, cy2, prob2 = trace['t2']['t2'], trace['t2']['a1'], trace['t2']['prob']
axes[0].scatter(cx1, cy1, c=prob, s=30)
axes[0].set_xlabel('a1')
axes[0].set_ylabel('t2')
axes[1].scatter(cx2, cy2, c=prob2, s=30)
axes[1].set_xlabel('t2')
axes[1].set_ylabel('a1')
plt.show()
which shows the trace of values:
As an alternative/complement to the confidence intervals, the Minimizer.emcee()
method uses Markov Chain Monte Carlo to sample the posterior probability distribution.
These distributions demonstrate the range of solutions that the data supports and we
refer to Minimizer.emcee() - calculating the posterior probability distribution of parameters where this methodology was used on the same problem.
Credible intervals (the Bayesian equivalent of the frequentist confidence interval) can be obtained with this method. MCMC can be used for model selection, to determine outliers, to marginalize over nuisance parameters, etcetera. For example, you may have fractionally underestimated the uncertainties on a dataset. MCMC can be used to estimate the true level of uncertainty on each data point. A tutorial on the possibilities offered by MCMC can be found at [1].
Confidence Interval Functions¶
- conf_interval(minimizer, result, p_names=None, sigmas=None, trace=False, maxiter=200, verbose=False, prob_func=None, min_rel_change=1e-05)¶
Calculate the confidence interval (CI) for parameters.
The parameter for which the CI is calculated will be varied, while the remaining parameters are re-optimized to minimize the chi-square. The resulting chi-square is used to calculate the probability with a given statistic (e.g., F-test). This function uses a 1d-rootfinder from SciPy to find the values resulting in the searched confidence region.
- Parameters:
minimizer (Minimizer) – The minimizer to use, holding objective function.
result (MinimizerResult) – The result of running minimize().
p_names (list, optional) – Names of the parameters for which the CI is calculated. If None (default), the CI is calculated for every parameter.
sigmas (list, optional) – The sigma-levels to find (default is [1, 2, 3]). See Notes below.
trace (bool, optional) – Defaults to False; if True, each result of a probability calculation is saved along with the parameter. This can be used to plot so-called “profile traces”.
maxiter (int, optional) – Maximum of iteration to find an upper limit (default is 200).
verbose (bool, optional) – Print extra debugging information (default is False).
prob_func (None or callable, optional) – Function to calculate the probability from the optimized chi-square. Default is None and uses the built-in function f_compare (i.e., F-test).
min_rel_change (float, optional) – Minimum relative change in probability (default is 1e-5).
- Returns:
output (dict) – A dictionary containing a list of
(sigma, vals)-tuples for each parameter.trace_dict (dict, optional) – Only if trace is True. Is a dictionary, the key is the parameter which was fixed. The values are again a dict with the names as keys, but with an additional key ‘prob’. Each contains an array of the corresponding values.
See also
Notes
The values for sigma are taken as the number of standard deviations for a normal distribution and converted to probabilities. That is, the default
sigma=[1, 2, 3]will use probabilities of 0.6827, 0.9545, and 0.9973. If any of the sigma values is less than 1, that will be interpreted as a probability. That is, a value of 1 and 0.6827 will give the same results, within precision.Examples
>>> from lmfit.printfuncs import * >>> mini = minimize(some_func, params) >>> mini.leastsq() True >>> report_errors(params) ... #report >>> ci = conf_interval(mini) >>> report_ci(ci) ... #report
Now with quantiles for the sigmas and using the trace.
>>> ci, trace = conf_interval(mini, sigmas=[0.5, 1, 2, 3], trace=True) >>> fixed = trace['para1']['para1'] >>> free = trace['para1']['not_para1'] >>> prob = trace['para1']['prob']
This makes it possible to plot the dependence between free and fixed parameters.
- conf_interval2d(minimizer, result, x_name, y_name, nx=10, ny=10, limits=None, prob_func=None, nsigma=5, chi2_out=False)¶
Calculate confidence regions for two fixed parameters.
The method itself is explained in conf_interval: here we are fixing two parameters.
- Parameters:
minimizer (Minimizer) – The minimizer to use, holding objective function.
result (MinimizerResult) – The result of running minimize().
x_name (str) – The name of the parameter which will be the x direction.
y_name (str) – The name of the parameter which will be the y direction.
nx (int, optional) – Number of points in the x direction (default is 10).
ny (int, optional) – Number of points in the y direction (default is 10).
limits (tuple, optional) – Should have the form
((x_upper, x_lower), (y_upper, y_lower)). If not given, the default is nsigma*stderr in each direction.prob_func (None or callable, deprecated) – Starting with version 1.2, this argument is unused and has no effect.
nsigma (float or int, optional) – Multiplier of stderr for limits (default is 5).
chi2_out (bool) – Whether to return chi-square at each coordinate instead of probability.
- Returns:
x (numpy.ndarray) – X-coordinates (same shape as nx).
y (numpy.ndarray) – Y-coordinates (same shape as ny).
grid (numpy.ndarray) – 2-D array (with shape
(nx, ny)) containing the calculated probabilities or chi-square.
See also
Examples
>>> mini = Minimizer(some_func, params) >>> result = mini.leastsq() >>> x, y, gr = conf_interval2d(mini, result, 'para1','para2') >>> plt.contour(x,y,gr)
- ci_report(ci, with_offset=True, ndigits=5)¶
Return text of a report for confidence intervals.
- Parameters:
- Returns:
Text of formatted report on confidence intervals.
- Return type:
