Note
Go to the end to download the full example code.
Fit Using Inequality Constraint¶
Sometimes specifying boundaries using min and max are not sufficient,
and more complicated (inequality) constraints are needed. In the example below
the center of the Lorentzian peak is constrained to be between 0-5 away from
the center of the Gaussian peak.
See also: https://lmfit.github.io/lmfit-py/constraints.html#using-inequality-constraints
import matplotlib.pyplot as plt
import numpy as np
from lmfit import Minimizer, create_params, report_fit
from lmfit.lineshapes import gaussian, lorentzian
def residual(pars, x, data):
model = (gaussian(x, pars['amp_g'], pars['cen_g'], pars['wid_g']) +
lorentzian(x, pars['amp_l'], pars['cen_l'], pars['wid_l']))
return model - data
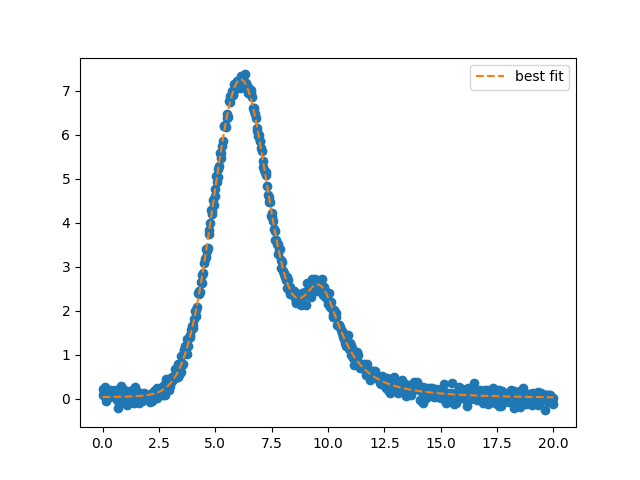
Generate the simulated data using a Gaussian and Lorentzian lineshape:
np.random.seed(0)
x = np.linspace(0, 20.0, 601)
data = (gaussian(x, 21, 6.1, 1.2) + lorentzian(x, 10, 9.6, 1.3) +
np.random.normal(scale=0.1, size=x.size))
Create the fitting parameters and set an inequality constraint for cen_l.
First, we add a new fitting parameter peak_split, which can take values
between 0 and 5. Afterwards, we constrain the value for cen_l using the
expression to be 'peak_split+cen_g':
pfit = create_params(amp_g=10, cen_g=5, wid_g=1, amp_l=10,
peak_split=dict(value=2.5, min=0, max=5),
cen_l=dict(expr='peak_split+cen_g'),
wid_l=dict(expr='wid_g'))
mini = Minimizer(residual, pfit, fcn_args=(x, data))
out = mini.leastsq()
best_fit = data + out.residual
Performing a fit, here using the leastsq algorithm, gives the following
fitting results:
report_fit(out.params)
[[Variables]]
amp_g: 21.2722842 +/- 0.05138772 (0.24%) (init = 10)
cen_g: 6.10496396 +/- 0.00334613 (0.05%) (init = 5)
wid_g: 1.21434954 +/- 0.00327317 (0.27%) (init = 1)
amp_l: 9.46504173 +/- 0.05445415 (0.58%) (init = 10)
peak_split: 3.52163544 +/- 0.01004618 (0.29%) (init = 2.5)
cen_l: 9.62659940 +/- 0.01066172 (0.11%) == 'peak_split+cen_g'
wid_l: 1.21434954 +/- 0.00327317 (0.27%) == 'wid_g'
[[Correlations]] (unreported correlations are < 0.100)
C(amp_g, wid_g) = +0.6199
C(amp_g, peak_split) = +0.3796
C(wid_g, peak_split) = +0.3445
C(amp_g, amp_l) = -0.2951
C(cen_g, amp_l) = -0.2761
C(amp_g, cen_g) = +0.1936
C(wid_g, amp_l) = -0.1651
C(cen_g, wid_g) = +0.1546
and figure:
Total running time of the script: (0 minutes 0.308 seconds)
