Note
Go to the end to download the full example code.
Fit with Algebraic Constraint¶
Example on how to use algebraic constraints using the expr attribute.
import matplotlib.pyplot as plt
from numpy import linspace, random
from lmfit.lineshapes import gaussian, lorentzian
from lmfit.models import GaussianModel, LinearModel, LorentzianModel
random.seed(0)
x = linspace(0.0, 20.0, 601)
data = (gaussian(x, amplitude=21, center=8.1, sigma=1.2) +
lorentzian(x, amplitude=10, center=9.6, sigma=2.4) +
0.01 + x*0.05 + random.normal(scale=0.23, size=x.size))
model = GaussianModel(prefix='g_') + LorentzianModel(prefix='l_') + LinearModel(prefix='line_')
params = model.make_params(g_amplitude=10, g_center=9, g_sigma=1,
line_slope=0, line_intercept=0)
params.add(name='total_amplitude', value=20)
params.set(l_amplitude=dict(expr='total_amplitude - g_amplitude'))
params.set(l_center=dict(expr='1.5+g_center'))
params.set(l_sigma=dict(expr='2*g_sigma'))
data_uncertainty = 0.021 # estimate of data error (for all data points)
init = model.eval(params, x=x)
result = model.fit(data, params, x=x, weights=1.0/data_uncertainty)
print(result.fit_report())
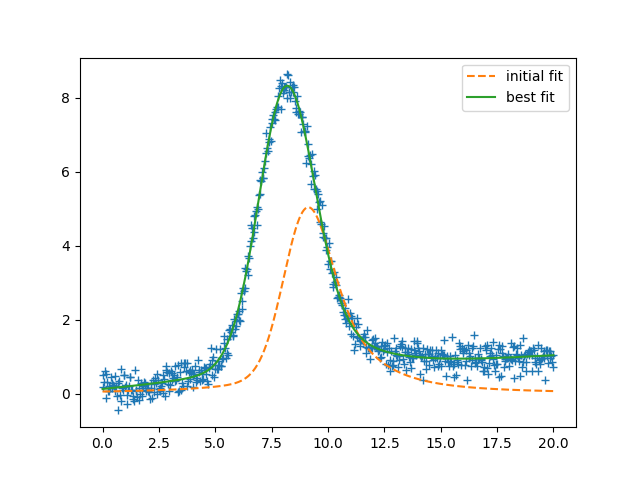
plt.plot(x, data, '+')
plt.plot(x, init, '--', label='initial fit')
plt.plot(x, result.best_fit, '-', label='best fit')
plt.legend()
plt.show()
[[Model]]
((Model(gaussian, prefix='g_') + Model(lorentzian, prefix='l_')) + Model(linear, prefix='line_'))
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 65
# data points = 601
# variables = 6
chi-square = 71878.3055
reduced chi-square = 120.803875
Akaike info crit = 2887.26503
Bayesian info crit = 2913.65660
R-squared = 0.98977025
[[Variables]]
g_amplitude: 21.1877635 +/- 0.32192128 (1.52%) (init = 10)
g_center: 8.11125903 +/- 0.01162987 (0.14%) (init = 9)
g_sigma: 1.20925819 +/- 0.01170853 (0.97%) (init = 1)
l_amplitude: 9.41261441 +/- 0.61672968 (6.55%) == 'total_amplitude - g_amplitude'
l_center: 9.61125903 +/- 0.01162987 (0.12%) == '1.5+g_center'
l_sigma: 2.41851637 +/- 0.02341707 (0.97%) == '2*g_sigma'
line_slope: 0.04615727 +/- 0.00170178 (3.69%) (init = 0)
line_intercept: 0.05128584 +/- 0.02448063 (47.73%) (init = 0)
g_fwhm: 2.84758536 +/- 0.02757149 (0.97%) == '2.3548200*g_sigma'
g_height: 6.98998378 +/- 0.05837066 (0.84%) == '0.3989423*g_amplitude/max(1e-15, g_sigma)'
l_fwhm: 4.83703275 +/- 0.04683414 (0.97%) == '2.0000000*l_sigma'
l_height: 1.23882905 +/- 0.08992735 (7.26%) == '0.3183099*l_amplitude/max(1e-15, l_sigma)'
total_amplitude: 30.6003779 +/- 0.36481425 (1.19%) (init = 20)
[[Correlations]] (unreported correlations are < 0.100)
C(g_amplitude, g_sigma) = +0.8662
C(g_amplitude, g_center) = +0.7496
C(line_slope, line_intercept) = -0.7144
C(g_center, total_amplitude) = -0.6952
C(g_center, g_sigma) = +0.6227
C(g_amplitude, total_amplitude) = -0.6115
C(line_intercept, total_amplitude) = -0.5883
C(g_sigma, total_amplitude) = -0.4115
C(g_center, line_intercept) = +0.3868
C(g_amplitude, line_intercept) = +0.1834
C(g_amplitude, line_slope) = +0.1825
C(g_sigma, line_slope) = +0.1739
Total running time of the script: (0 minutes 0.332 seconds)
