Note
Go to the end to download the full example code.
Fit Multiple Data Sets Using Model Interface¶
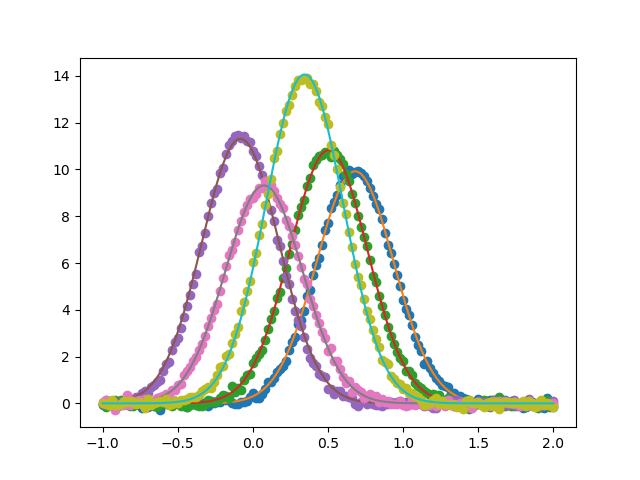
Fitting multiple (simulated) Gaussian data sets simultaneously, using the Model interface.
All minimizers require the residual array to be one-dimensional. Therefore,
in the objective function we need to flatten the array before
returning it.
import matplotlib.pyplot as plt
import numpy as np
from lmfit import Parameters, minimize, report_fit
from lmfit.models import GaussianModel
Create N simulated Gaussian data sets
N = 5
np.random.seed(2021)
x = np.linspace(-1, 2, 151)
data = []
for _ in np.arange(N):
params = Parameters()
params.add('amplitude', value=0.60 + 9.50*np.random.rand())
params.add('center', value=-0.20 + 1.20*np.random.rand())
params.add('sigma', value=0.25 + 0.03*np.random.rand())
dat = (GaussianModel().eval(x=x, params=params) +
np.random.normal(size=x.size, scale=0.1))
data.append(dat)
data = np.array(data)
The objective function will extract and evaluate a Gaussian from the compound model
def objective(params, x, data, model):
"""Calculate total residual for fits of Gaussians to several data sets."""
ndata, _ = data.shape
resid = 0.0*data[:]
# make residual per data set
for i in range(ndata):
components = model.components[i].eval(params=params, x=x)
resid[i, :] = data[i, :] - components
# now flatten this to a 1D array, as minimize() needs
return resid.flatten()
Create a composite model by adding Gaussians
Prepare the fitting parameters and constrain n2_sigma, …, nN_sigma to be equal to n1_sigma
fit_params = model.make_params()
for iy, y in enumerate(data):
fit_params.add(f'n{iy+1}_amplitude', value=0.5, min=0.0, max=200)
fit_params.add(f'n{iy+1}_center', value=0.4, min=-2.0, max=2.0)
fit_params.add(f'n{iy+1}_sigma', value=0.3, min=0.01, max=3.0)
if iy > 0:
fit_params[f'n{iy+1}_sigma'].expr = 'n1_sigma'
Run the global fit and show the fitting result
[[Variables]]
n1_amplitude: 6.40552155 +/- 0.01575090 (0.25%) (init = 0.5)
n1_center: 0.68032886 +/- 8.6209e-04 (0.13%) (init = 0.4)
n1_sigma: 0.25743367 +/- 3.4131e-04 (0.13%) (init = 0.3)
n2_amplitude: 6.98975626 +/- 0.01585971 (0.23%) (init = 0.5)
n2_center: 0.50446887 +/- 7.9004e-04 (0.16%) (init = 0.4)
n2_sigma: 0.25743367 +/- 3.4131e-04 (0.13%) == 'n1_sigma'
n3_amplitude: 7.30195609 +/- 0.01592142 (0.22%) (init = 0.5)
n3_center: -0.08261146 +/- 7.5626e-04 (0.92%) (init = 0.4)
n3_sigma: 0.25743367 +/- 3.4131e-04 (0.13%) == 'n1_sigma'
n4_amplitude: 6.01461340 +/- 0.01568302 (0.26%) (init = 0.5)
n4_center: 0.07382999 +/- 9.1812e-04 (1.24%) (init = 0.4)
n4_sigma: 0.25743367 +/- 3.4131e-04 (0.13%) == 'n1_sigma'
n5_amplitude: 9.07744386 +/- 0.01631785 (0.18%) (init = 0.5)
n5_center: 0.34402084 +/- 6.0834e-04 (0.18%) (init = 0.4)
n5_sigma: 0.25743367 +/- 3.4131e-04 (0.13%) == 'n1_sigma'
n1_fwhm: 0.60620995 +/- 8.0373e-04 (0.13%) == '2.3548200*n1_sigma'
n1_height: 9.92657076 +/- 0.02440907 (0.25%) == '0.3989423*n1_amplitude/max(1e-15, n1_sigma)'
n2_fwhm: 0.60620995 +/- 8.0373e-04 (0.13%) == '2.3548200*n2_sigma'
n2_height: 10.8319533 +/- 0.02457770 (0.23%) == '0.3989423*n2_amplitude/max(1e-15, n2_sigma)'
n3_fwhm: 0.60620995 +/- 8.0373e-04 (0.13%) == '2.3548200*n3_sigma'
n3_height: 11.3157661 +/- 0.02467331 (0.22%) == '0.3989423*n3_amplitude/max(1e-15, n3_sigma)'
n4_fwhm: 0.60620995 +/- 8.0373e-04 (0.13%) == '2.3548200*n4_sigma'
n4_height: 9.32078443 +/- 0.02430386 (0.26%) == '0.3989423*n4_amplitude/max(1e-15, n4_sigma)'
n5_fwhm: 0.60620995 +/- 8.0373e-04 (0.13%) == '2.3548200*n5_sigma'
n5_height: 14.0672212 +/- 0.02528769 (0.18%) == '0.3989423*n5_amplitude/max(1e-15, n5_sigma)'
[[Correlations]] (unreported correlations are < 0.100)
C(n1_sigma, n5_amplitude) = +0.3688
C(n1_sigma, n3_amplitude) = +0.3040
C(n1_sigma, n2_amplitude) = +0.2922
C(n1_amplitude, n1_sigma) = +0.2696
C(n1_sigma, n4_amplitude) = +0.2542
C(n3_amplitude, n5_amplitude) = +0.1121
C(n2_amplitude, n5_amplitude) = +0.1077
Plot the data sets and fits
Total running time of the script: (0 minutes 0.414 seconds)
