Note
Go to the end to download the full example code.
Fit using the Model interface¶
This notebook shows a simple example of using the lmfit.Model class. For
more information please refer to:
https://lmfit.github.io/lmfit-py/model.html#the-model-class.
import numpy as np
from pandas import Series
from lmfit import Model, Parameter, report_fit
The Model class is a flexible, concise curve fitter. I will illustrate
fitting example data to an exponential decay.
The parameters are in no particular order. We’ll need some example data. I
will use N=7 and tau=3, and add a little noise.
t = np.linspace(0, 5, num=1000)
np.random.seed(2021)
data = decay(t, 7, 3) + np.random.randn(t.size)
Simplest Usage
The Model infers the parameter names by inspecting the arguments of the
function, decay. Then I passed the independent variable, t, and
initial guesses for each parameter. A residual function is automatically
defined, and a least-squared regression is performed.
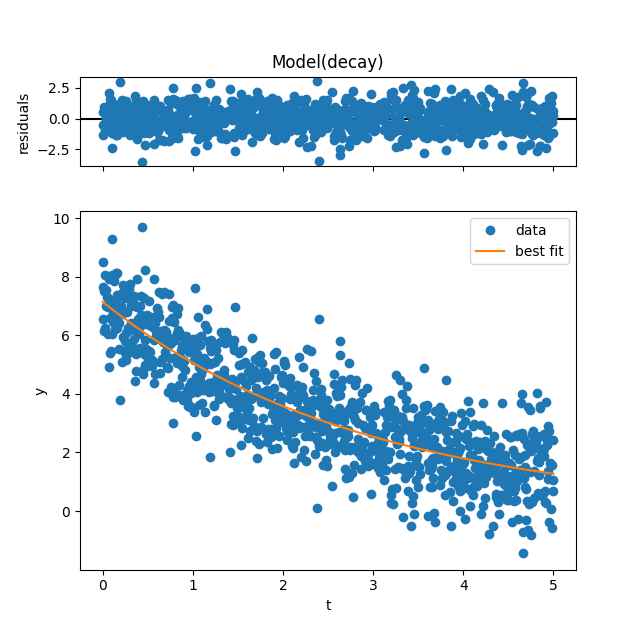
We can immediately see the best-fit values:
print(result.values)
{'N': 7.146693193035106, 'tau': 2.898028980706992}
and use these best-fit parameters for plotting with the plot function:
result.plot()
We can review the best-fit Parameters by accessing result.params:
result.params.pretty_print()
Name Value Min Max Stderr Vary Expr Brute_Step
N 7.147 -inf inf 0.0913 True None None
tau 2.898 -inf inf 0.06299 True None None
More information about the fit is stored in the result, which is an
lmfit.MimimizerResult object (see:
https://lmfit.github.io/lmfit-py/fitting.html#lmfit.minimizer.MinimizerResult)
Specifying Bounds and Holding Parameters Constant
Above, the Model class implicitly builds Parameter objects from
keyword arguments of fit that match the arguments of decay. You can
build the Parameter objects explicitly; the following is equivalent.
[[Variables]]
N: 7.14669319 +/- 0.09130428 (1.28%) (init = 10)
tau: 2.89802898 +/- 0.06299118 (2.17%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.7533
By building Parameter objects explicitly, you can specify bounds
(min, max) and set parameters constant (vary=False).
[[Variables]]
N: 7 (fixed)
tau: 2.97663118 +/- 0.04347476 (1.46%) (init = 1)
Defining Parameters in Advance
Passing parameters to fit can become unwieldy. As an alternative, you
can extract the parameters from model like so, set them individually,
and pass them to fit.
[[Variables]]
N: 7.14669316 +/- 0.09130423 (1.28%) (init = 10)
tau: 2.89802901 +/- 0.06299127 (2.17%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.7533
Keyword arguments override params, resetting value and all other
properties (min, max, vary).
[[Variables]]
N: 7.14669316 +/- 0.09130423 (1.28%) (init = 10)
tau: 2.89802901 +/- 0.06299127 (2.17%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.7533
The input parameters are not modified by fit. They can be reused,
retaining the same initial value. If you want to use the result of one fit
as the initial guess for the next, simply pass params=result.params.
#TODO/FIXME: not sure if there ever way a “helpful exception”, but currently
#it raises a ValueError: The input contains nan values.
#*A Helpful Exception*
#All this implicit magic makes it very easy for the user to neglect to set a
#parameter. The fit function checks for this and raises a helpful exception.
# #result = model.fit(data, t=t, tau=1) # N unspecified
#An extra parameter that cannot be matched to the model function will
#throw a UserWarning, but it will not raise, leaving open the possibility
#of unforeseen extensions calling for some parameters.
Weighted Fits
Use the sigma argument to perform a weighted fit. If you prefer to think
of the fit in term of weights, sigma=1/weights.
[[Variables]]
N: 6.98535179 +/- 0.28002384 (4.01%) (init = 10)
tau: 2.97268236 +/- 0.11134755 (3.75%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.9311
Handling Missing Data
By default, attempting to fit data that includes a NaN, which
conventionally indicates a “missing” observation, raises a lengthy exception.
You can choose to omit (i.e., skip over) missing values instead.
data_with_holes = data.copy()
data_with_holes[[5, 500, 700]] = np.nan # Replace arbitrary values with NaN.
model = Model(decay, independent_vars=['t'], nan_policy='omit')
result = model.fit(data_with_holes, params, t=t)
report_fit(result.params)
[[Variables]]
N: 7.15448795 +/- 0.09181809 (1.28%) (init = 10)
tau: 2.89285089 +/- 0.06306004 (2.18%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.7542
If you don’t want to ignore missing values, you can set the model to raise proactively, checking for missing values before attempting the fit.
Uncomment to see the error #model = Model(decay, independent_vars=[‘t’], nan_policy=’raise’) #result = model.fit(data_with_holes, params, t=t)
The default setting is nan_policy='raise', which does check for NaNs and
raises an exception when present.
Null-checking relies on pandas.isnull if it is available. If pandas
cannot be imported, it silently falls back on numpy.isnan.
Data Alignment
Imagine a collection of time series data with different lengths. It would be
convenient to define one sufficiently long array t and use it for each
time series, regardless of length. pandas
(https://pandas.pydata.org/pandas-docs/stable/) provides tools for aligning
indexed data. And, unlike most wrappers to scipy.leastsq, Model can
handle pandas objects out of the box, using its data alignment features.
Here I take just a slice of the data and fit it to the full t. It is
automatically aligned to the correct section of t using Series’ index.
model = Model(decay, independent_vars=['t'])
truncated_data = Series(data)[200:800] # data points 200-800
t = Series(t) # all 1000 points
result = model.fit(truncated_data, params, t=t)
report_fit(result.params)
[[Variables]]
N: 7.10725864 +/- 0.24259071 (3.41%) (init = 10)
tau: 2.92503564 +/- 0.13481789 (4.61%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.9320
Data with missing entries and an unequal length still aligns properly.
model = Model(decay, independent_vars=['t'], nan_policy='omit')
truncated_data_with_holes = Series(data_with_holes)[200:800]
result = model.fit(truncated_data_with_holes, params, t=t)
report_fit(result.params)
[[Variables]]
N: 7.11270194 +/- 0.24334895 (3.42%) (init = 10)
tau: 2.92065227 +/- 0.13488230 (4.62%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(N, tau) = -0.9320
Total running time of the script: (0 minutes 0.485 seconds)
