Note
Go to the end to download the full example code.
Fit comparing leastsq and basin hopping, or other methods¶
This example compares the leastsq and basinhopping algorithms
on a decaying sine wave. Note that this can be used to compare other
fitting algorithms too.
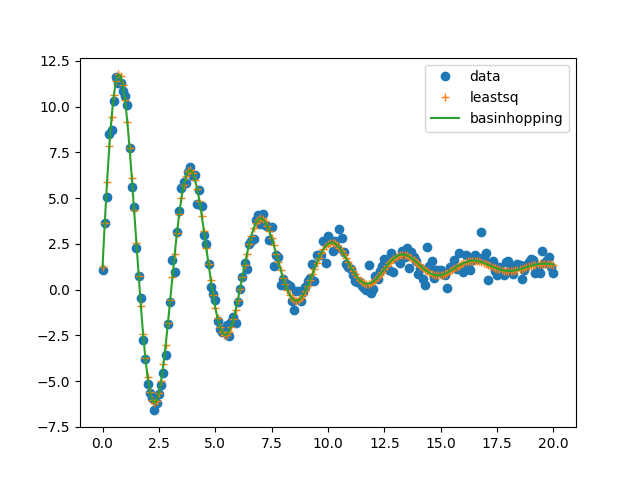
# Fit using leastsq:
[[Model]]
Model(sine_decay)
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 31
# data points = 201
# variables = 4
chi-square = 37.4526504
reduced chi-square = 0.19011498
Akaike info crit = -329.725713
Bayesian info crit = -316.512493
R-squared = 0.97868751
[[Variables]]
amplitude: 12.4411385 +/- 0.18965087 (1.52%) (init = 10)
frequency: 1.99852115 +/- 0.00324489 (0.16%) (init = 2)
decay: 4.54866058 +/- 0.09723868 (2.14%) (init = 2)
offset: 1.25370110 +/- 0.03107114 (2.48%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amplitude, decay) = -0.7241
C(amplitude, offset) = -0.1418
#####################
# Fit using basinhopping:
[[Model]]
Model(sine_decay)
[[Fit Statistics]]
# fitting method = basinhopping
# function evals = 24867
# data points = 201
# variables = 4
chi-square = 37.4526504
reduced chi-square = 0.19011498
Akaike info crit = -329.725713
Bayesian info crit = -316.512493
R-squared = 0.97868751
[[Variables]]
amplitude: 12.4411365 +/- 0.18862101 (1.52%) (init = 10)
frequency: 1.99852108 +/- 0.00326218 (0.16%) (init = 2)
decay: 4.54866141 +/- 0.09621984 (2.12%) (init = 2)
offset: 1.25370114 +/- 0.03106851 (2.48%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amplitude, decay) = -0.7205
C(amplitude, offset) = -0.1412
import sys
import matplotlib.pyplot as plt
import numpy as np
import lmfit
VALID_METHODS = ['least_squares', 'differential_evolution', 'brute',
'basinhopping', 'ampgo', 'nelder', 'lbfgsb', 'powell', 'cg',
'newton', 'cobyla', 'bfgs', 'tnc', 'trust-ncg', 'trust-exact',
'trust-krylov', 'trust-constr', 'dogleg', 'slsqp', 'emcee',
'shgo', 'dual_annealing']
def sine_decay(x, amplitude, frequency, decay, offset):
return offset + amplitude * np.sin(x*frequency) * np.exp(-x/decay)
x = np.linspace(0, 20, 201)
np.random.seed(2)
ydat = sine_decay(x, 12.5, 2.0, 4.5, 1.25) + np.random.normal(size=len(x), scale=0.40)
model = lmfit.Model(sine_decay)
params = model.make_params(amplitude={'value': 10, 'min': 0, 'max': 1000},
frequency={'value': 2.0, 'min': 0, 'max': 6.0},
decay={'value': 2.0, 'min': 0.001, 'max': 12},
offset=1.0)
# fit with leastsq
result0 = model.fit(ydat, params, x=x, method='leastsq')
print("# Fit using leastsq:")
print(result0.fit_report())
method2 = 'basinhopping'
if len(sys.argv) > 1 and sys.argv[1] in VALID_METHODS:
method2 = sys.argv[1]
# fit with other method
result = model.fit(ydat, params, x=x, method=method2)
print(f"\n#####################\n# Fit using {method2}:")
print(result.fit_report())
# plot comparison
plt.plot(x, ydat, 'o', label='data')
plt.plot(x, result0.best_fit, '+', label='leastsq')
plt.plot(x, result.best_fit, '-', label=method2)
plt.legend()
plt.show()
Total running time of the script: (0 minutes 3.016 seconds)
