Note
Go to the end to download the full example code.
Confidence - chi2 maps¶
# <examples/doc_confidence_chi2_maps.py>
import matplotlib.pyplot as plt
import numpy as np
from lmfit import conf_interval, conf_interval2d, report_ci
from lmfit.lineshapes import gaussian
from lmfit.models import GaussianModel, LinearModel
sigma_levels = [1, 2, 3]
rng = np.random.default_rng(seed=102)
set up data – deliberately adding imperfections and a small amount of non-Gaussian noise
npts = 501
x = np.linspace(1, 100, num=npts)
noise = rng.normal(scale=0.3, size=npts) + 0.2*rng.f(3, 9, size=npts)
y = (gaussian(x, amplitude=83, center=47., sigma=5.)
+ 0.02*x + 4 + 0.25*np.cos((x-20)/8.0) + noise)
mod = GaussianModel() + LinearModel()
params = mod.make_params(amplitude=100, center=50, sigma=5,
slope=0, intecept=2)
out = mod.fit(y, params, x=x)
print(out.fit_report(correl_mode='table'))
[[Model]]
(Model(gaussian) + Model(linear))
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 31
# data points = 501
# variables = 5
chi-square = 103.861381
reduced chi-square = 0.20939794
Akaike info crit = -778.348033
Bayesian info crit = -757.265003
R-squared = 0.93782756
[[Variables]]
amplitude: 78.8171374 +/- 1.21910939 (1.55%) (init = 100)
center: 47.0751649 +/- 0.07576660 (0.16%) (init = 50)
sigma: 4.93298753 +/- 0.07984021 (1.62%) (init = 5)
slope: 0.01839006 +/- 7.1957e-04 (3.91%) (init = 0)
intercept: 4.39234411 +/- 0.04420227 (1.01%) (init = 0)
fwhm: 11.6162977 +/- 0.18800933 (1.62%) == '2.3548200*sigma'
height: 6.37412722 +/- 0.08603873 (1.35%) == '0.3989423*amplitude/max(1e-15, sigma)'
[[Correlations]]
+-----------+-----------+-----------+-----------+-----------+-----------+
| Variable | amplitude | center | sigma | slope | intercept |
+-----------+-----------+-----------+-----------+-----------+-----------+
| amplitude | +1.0000 | -0.0074 | +0.6371 | +0.0721 | -0.3373 |
| center | -0.0074 | +1.0000 | -0.0048 | -0.1026 | +0.0864 |
| sigma | +0.6371 | -0.0048 | +1.0000 | +0.0459 | -0.2149 |
| slope | +0.0721 | -0.1026 | +0.0459 | +1.0000 | -0.8421 |
| intercept | -0.3373 | +0.0864 | -0.2149 | -0.8421 | +1.0000 |
+-----------+-----------+-----------+-----------+-----------+-----------+
run conf_intervale, print report
ci = conf_interval(out, out, sigmas=sigma_levels)
print("## Confidence Report:")
report_ci(ci)
## Confidence Report:
99.73% 95.45% 68.27% _BEST_ 68.27% 95.45% 99.73%
amplitude: -3.62610 -2.41983 -1.21237 78.81714 +1.22111 +2.45479 +3.70515
center : -0.22849 -0.15214 -0.07584 47.07516 +0.07587 +0.15225 +0.22873
sigma : -0.23335 -0.15640 -0.07870 4.93299 +0.08000 +0.16158 +0.24509
slope : -0.00217 -0.00144 -0.00072 0.01839 +0.00072 +0.00144 +0.00217
intercept: -0.13326 -0.08860 -0.04423 4.39234 +0.04421 +0.08854 +0.13312
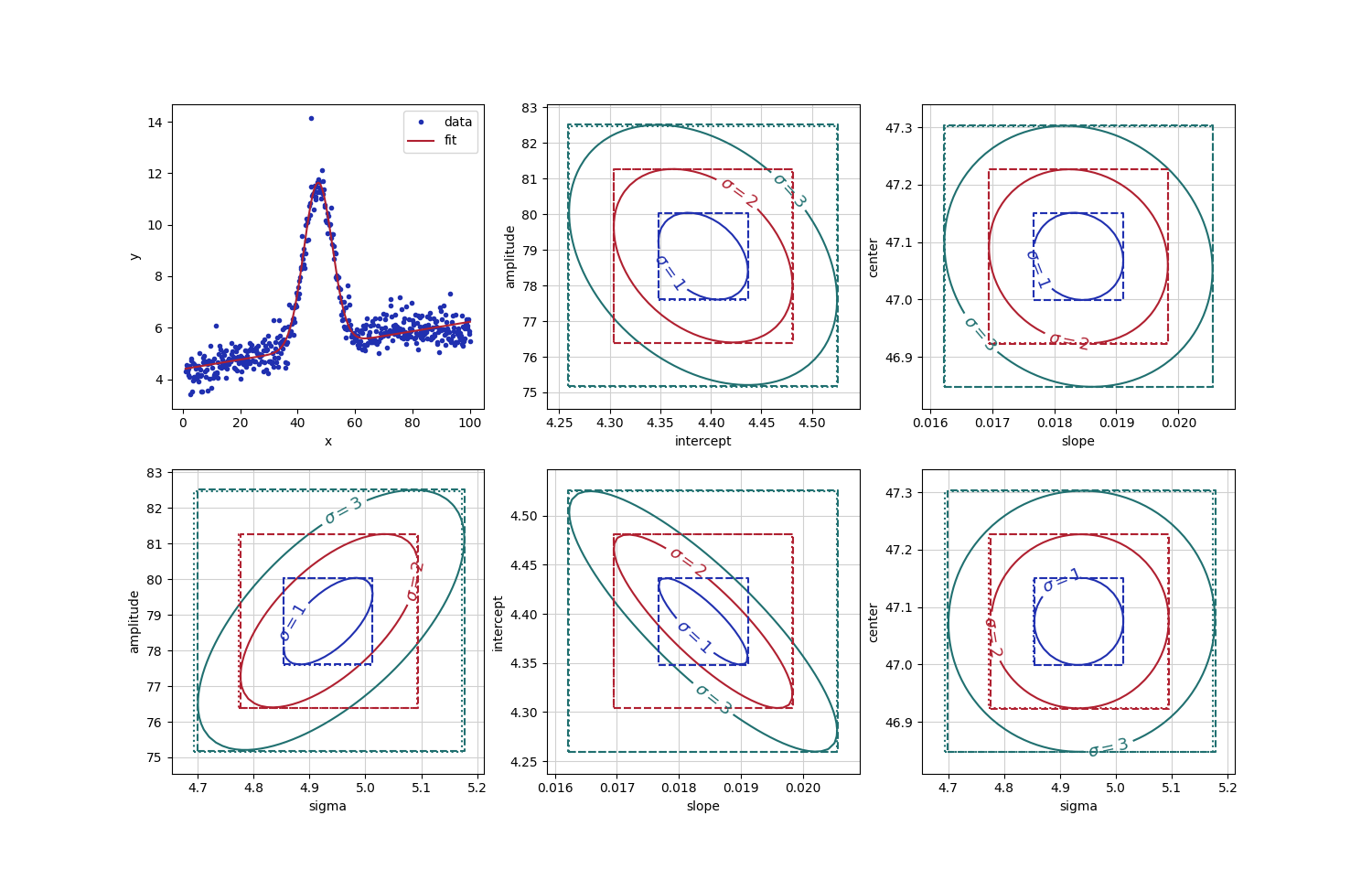
plot initial fit
colors = ('#2030b0', '#b02030', '#207070')
fig, axes = plt.subplots(2, 3, figsize=(15, 9.5))
axes[0, 0].plot(x, y, 'o', markersize=3, label='data', color=colors[0])
axes[0, 0].plot(x, out.best_fit, label='fit', color=colors[1])
axes[0, 0].set_xlabel('x')
axes[0, 0].set_ylabel('y')
axes[0, 0].legend()
aix, aiy = 0, 0
nsamples = 50
explicitly_calculate_sigma = True
for pairs in (('sigma', 'amplitude'), ('intercept', 'amplitude'),
('slope', 'intercept'), ('slope', 'center'), ('sigma', 'center')):
xpar, ypar = pairs
if explicitly_calculate_sigma:
print(f"Generating chi-square map for {pairs}")
c_x, c_y, chi2_mat = conf_interval2d(out, out, xpar, ypar,
nsamples, nsamples, nsigma=3.5,
chi2_out=True)
# explicitly calculate sigma matrix: sigma increases chi_square
# from chi_square_best
# to chi_square + sigma**2 * reduced_chi_square
# so: sigma = sqrt((chi2-chi2_best)/ reduced_chi_square)
chi2_min = chi2_mat.min()
sigma_mat = np.sqrt((chi2_mat-chi2_min)/out.redchi)
else:
print(f"Generating sigma map for {pairs}")
# or, you could just calculate the matrix of probabilities as:
c_x, c_y, sigma_mat = conf_interval2d(out, out, xpar, ypar,
nsamples, nsamples, nsigma=3.5)
aix += 1
if aix == 2:
aix = 0
aiy += 1
ax = axes[aix, aiy]
cnt = ax.contour(c_x, c_y, sigma_mat, levels=sigma_levels, colors=colors,
linestyles='-')
ax.clabel(cnt, inline=True, fmt=r"$\sigma=%.0f$", fontsize=13)
# draw boxes for estimated uncertaties:
# dotted : scaled stderr from initial fit
# dashed : values found from conf_interval()
xv = out.params[xpar].value
xs = out.params[xpar].stderr
yv = out.params[ypar].value
ys = out.params[ypar].stderr
cix = ci[xpar]
ciy = ci[ypar]
nc = len(sigma_levels)
for i in sigma_levels:
# dotted line: scaled stderr
ax.plot((xv-i*xs, xv+i*xs, xv+i*xs, xv-i*xs, xv-i*xs),
(yv-i*ys, yv-i*ys, yv+i*ys, yv+i*ys, yv-i*ys),
linestyle='dotted', color=colors[i-1])
# dashed line: refined uncertainties from conf_interval
xsp, xsm = cix[nc+i][1], cix[nc-i][1]
ysp, ysm = ciy[nc+i][1], ciy[nc-i][1]
ax.plot((xsm, xsp, xsp, xsm, xsm), (ysm, ysm, ysp, ysp, ysm),
linestyle='dashed', color=colors[i-1])
ax.set_xlabel(xpar)
ax.set_ylabel(ypar)
ax.grid(True, color='#d0d0d0')
plt.show()
# <end examples/doc_confidence_chi2_maps.py>
Generating chi-square map for ('sigma', 'amplitude')
Generating chi-square map for ('intercept', 'amplitude')
Generating chi-square map for ('slope', 'intercept')
Generating chi-square map for ('slope', 'center')
Generating chi-square map for ('sigma', 'center')
Total running time of the script: (0 minutes 46.032 seconds)
