Note
Go to the end to download the full example code.
Calculate Confidence Intervals¶
Define the residual function, specify “true” parameter values, and generate a synthetic data set with some noise:
def residual(pars, x, data=None):
argu = (x*pars['decay'])**2
shift = pars['shift']
if abs(shift) > pi/2:
shift = shift - sign(shift)*pi
model = pars['amp']*sin(shift + x/pars['period']) * exp(-argu)
if data is None:
return model
return model - data
p_true = create_params(amp=14.0, period=5.33, shift=0.123, decay=0.010)
x = linspace(0.0, 250.0, 2500)
random.seed(2021)
noise = random.normal(scale=0.7215, size=x.size)
data = residual(p_true, x) + noise
Create fitting parameters and set initial values:
fit_params = create_params(amp=13.0, period=2, shift=0.0, decay=0.020)
Set-up the minimizer and perform the fit using leastsq algorithm, and
show the report:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 95
# data points = 2500
# variables = 4
chi-square = 1277.24638
reduced chi-square = 0.51171730
Akaike info crit = -1670.96059
Bayesian info crit = -1647.66441
[[Variables]]
amp: 14.0708269 +/- 0.04936878 (0.35%) (init = 13)
period: 5.32980958 +/- 0.00273143 (0.05%) (init = 2)
shift: 0.12156317 +/- 0.00482312 (3.97%) (init = 0)
decay: 0.01002489 +/- 4.0726e-05 (0.41%) (init = 0.02)
[[Correlations]] (unreported correlations are < 0.100)
C(period, shift) = +0.8002
C(amp, decay) = +0.5758
Calculate the confidence intervals for parameters and display the results:
ci, tr = conf_interval(mini, out, trace=True)
report_ci(ci)
names = out.params.keys()
i = 0
gs = plt.GridSpec(4, 4)
sx = {}
sy = {}
for fixed in names:
j = 0
for free in names:
if j in sx and i in sy:
ax = plt.subplot(gs[i, j], sharex=sx[j], sharey=sy[i])
elif i in sy:
ax = plt.subplot(gs[i, j], sharey=sy[i])
sx[j] = ax
elif j in sx:
ax = plt.subplot(gs[i, j], sharex=sx[j])
sy[i] = ax
else:
ax = plt.subplot(gs[i, j])
sy[i] = ax
sx[j] = ax
if i < 3:
plt.setp(ax.get_xticklabels(), visible=False)
else:
ax.set_xlabel(free)
if j > 0:
plt.setp(ax.get_yticklabels(), visible=False)
else:
ax.set_ylabel(fixed)
res = tr[fixed]
prob = res['prob']
f = prob < 0.96
x, y = res[free], res[fixed]
ax.scatter(x[f], y[f], c=1-prob[f], s=25*(1-prob[f]+0.5))
ax.autoscale(1, 1)
j += 1
i += 1
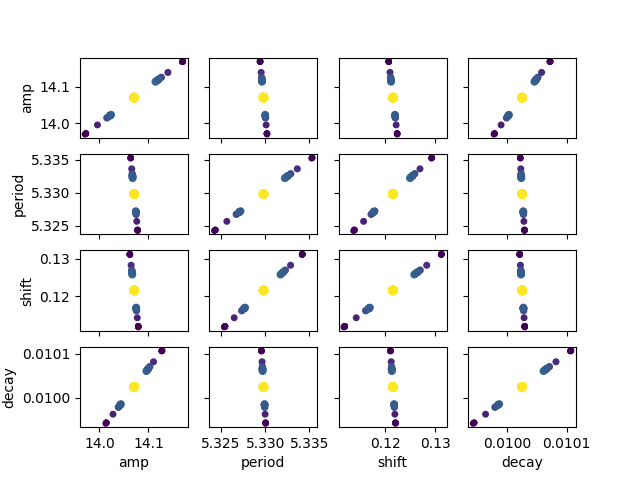
99.73% 95.45% 68.27% _BEST_ 68.27% 95.45% 99.73%
amp : -0.14795 -0.09863 -0.04934 14.07083 +0.04939 +0.09886 +0.14847
period: -0.00818 -0.00546 -0.00273 5.32981 +0.00273 +0.00548 +0.00822
shift : -0.01446 -0.00964 -0.00482 0.12156 +0.00482 +0.00965 +0.01449
decay : -0.00012 -0.00008 -0.00004 0.01002 +0.00004 +0.00008 +0.00012
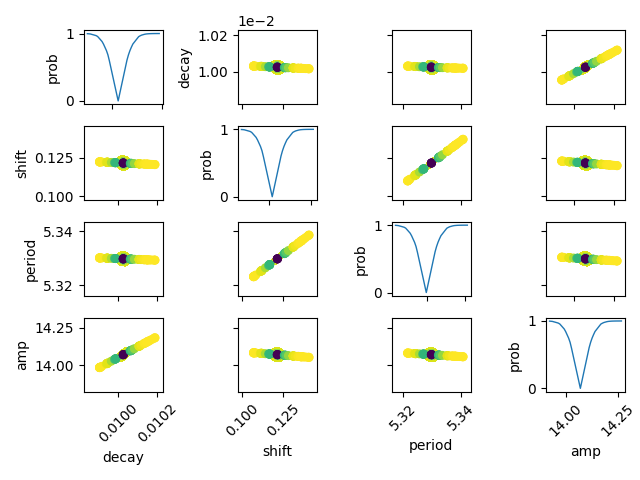
It is also possible to calculate the confidence regions for two fixed
parameters using the function conf_interval2d:
names = list(out.params.keys())
plt.figure()
for i in range(4):
for j in range(4):
indx = 16-j*4-i
ax = plt.subplot(4, 4, indx)
ax.ticklabel_format(style='sci', scilimits=(-2, 2), axis='y')
# set-up labels and tick marks
ax.tick_params(labelleft=False, labelbottom=False)
if indx in (2, 5, 9, 13):
plt.ylabel(names[j])
ax.tick_params(labelleft=True)
if indx == 1:
ax.tick_params(labelleft=True)
if indx in (13, 14, 15, 16):
plt.xlabel(names[i])
ax.tick_params(labelbottom=True)
[label.set_rotation(45) for label in ax.get_xticklabels()]
if i != j:
x, y, m = conf_interval2d(mini, out, names[i], names[j], 20, 20)
plt.contourf(x, y, m, linspace(0, 1, 10))
x = tr[names[i]][names[i]]
y = tr[names[i]][names[j]]
pr = tr[names[i]]['prob']
s = argsort(x)
plt.scatter(x[s], y[s], c=pr[s], s=30, lw=1)
else:
x = tr[names[i]][names[i]]
y = tr[names[i]]['prob']
t, s = unique(x, True)
f = interp1d(t, y[s], 'slinear')
xn = linspace(x.min(), x.max(), 50)
plt.plot(xn, f(xn), lw=1)
plt.ylabel('prob')
ax.tick_params(labelleft=True)
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 10.971 seconds)
