Note
Go to the end to download the full example code.
Global minimization using the brute method (a.k.a. grid search)¶
This notebook shows a simple example of using lmfit.minimize.brute that
uses the method with the same name from scipy.optimize.
The method computes the function’s value at each point of a multidimensional
grid of points, to find the global minimum of the function. It behaves
identically to scipy.optimize.brute in case finite bounds are given on
all varying parameters, but will also deal with non-bounded parameters
(see below).
import copy
from matplotlib.colors import LogNorm
import matplotlib.pyplot as plt
import numpy as np
from lmfit import Minimizer, create_params, fit_report
Let’s start with the example given in the documentation of SciPy:
“We illustrate the use of brute to seek the global minimum of a function of
two variables that is given as the sum of a positive-definite quadratic and
two deep “Gaussian-shaped” craters. Specifically, define the objective
function f as the sum of three other functions, f = f1 + f2 + f3. We
suppose each of these has a signature (z, *params), where z = (x, y),
and params and the functions are as defined below.”
First, we create a set of Parameters where all variables except x and
y are given fixed values.
Just as in the documentation we will do a grid search between -4 and
4 and use a stepsize of 0.25. The bounds can be set as usual with
the min and max attributes, and the stepsize is set using
brute_step.
params = create_params(a=dict(value=2, vary=False),
b=dict(value=3, vary=False),
c=dict(value=7, vary=False),
d=dict(value=8, vary=False),
e=dict(value=9, vary=False),
f=dict(value=10, vary=False),
g=dict(value=44, vary=False),
h=dict(value=-1, vary=False),
i=dict(value=2, vary=False),
j=dict(value=26, vary=False),
k=dict(value=1, vary=False),
l=dict(value=-2, vary=False),
scale=dict(value=0.5, vary=False),
x=dict(value=0.0, vary=True, min=-4, max=4, brute_step=0.25),
y=dict(value=0.0, vary=True, min=-4, max=4, brute_step=0.25))
Second, create the three functions and the objective function:
def f1(p):
par = p.valuesdict()
return (par['a'] * par['x']**2 + par['b'] * par['x'] * par['y'] +
par['c'] * par['y']**2 + par['d']*par['x'] + par['e']*par['y'] +
par['f'])
def f2(p):
par = p.valuesdict()
return (-1.0*par['g']*np.exp(-((par['x']-par['h'])**2 +
(par['y']-par['i'])**2) / par['scale']))
def f3(p):
par = p.valuesdict()
return (-1.0*par['j']*np.exp(-((par['x']-par['k'])**2 +
(par['y']-par['l'])**2) / par['scale']))
def f(params):
return f1(params) + f2(params) + f3(params)
Performing the actual grid search is done with:
fitter = Minimizer(f, params)
result = fitter.minimize(method='brute')
, which will increment x and y between -4 in increments of
0.25 until 4 (not inclusive).
grid_x, grid_y = (np.unique(par.ravel()) for par in result.brute_grid)
print(grid_x)
[-4. -3.75 -3.5 -3.25 -3. -2.75 -2.5 -2.25 -2. -1.75 -1.5 -1.25
-1. -0.75 -0.5 -0.25 0. 0.25 0.5 0.75 1. 1.25 1.5 1.75
2. 2.25 2.5 2.75 3. 3.25 3.5 3.75]
The objective function is evaluated on this grid, and the raw output from
scipy.optimize.brute is stored in the MinimizerResult as
brute_<parname> attributes. These attributes are:
result.brute_x0 – A 1-D array containing the coordinates of a point at
which the objective function had its minimum value.
print(result.brute_x0)
[-1. 1.75]
result.brute_fval – Function value at the point x0.
print(result.brute_fval)
-2.8923637137222027
result.brute_grid – Representation of the evaluation grid. It has the
same length as x0.
print(result.brute_grid)
[[[-4. -4. -4. ... -4. -4. -4. ]
[-3.75 -3.75 -3.75 ... -3.75 -3.75 -3.75]
[-3.5 -3.5 -3.5 ... -3.5 -3.5 -3.5 ]
...
[ 3.25 3.25 3.25 ... 3.25 3.25 3.25]
[ 3.5 3.5 3.5 ... 3.5 3.5 3.5 ]
[ 3.75 3.75 3.75 ... 3.75 3.75 3.75]]
[[-4. -3.75 -3.5 ... 3.25 3.5 3.75]
[-4. -3.75 -3.5 ... 3.25 3.5 3.75]
[-4. -3.75 -3.5 ... 3.25 3.5 3.75]
...
[-4. -3.75 -3.5 ... 3.25 3.5 3.75]
[-4. -3.75 -3.5 ... 3.25 3.5 3.75]
[-4. -3.75 -3.5 ... 3.25 3.5 3.75]]]
result.brute_Jout – Function values at each point of the evaluation
grid, i.e., Jout = func(*grid).
print(result.brute_Jout)
[[134. 119.6875 106.25 ... 74.18749997 85.24999999
97.1875 ]
[129.125 115. 101.75 ... 74.74999948 85.99999987
98.12499997]
[124.5 110.5625 97.5 ... 75.5624928 86.99999818
99.31249964]
...
[ 94.12499965 85.24999772 77.24998843 ... 192. 208.5
225.875 ]
[ 96.49999997 87.81249979 79.99999892 ... 199.8125 216.5
234.0625 ]
[ 99.125 90.62499998 82.99999992 ... 207.875 224.75
242.5 ]]
Reassuringly, the obtained results are identical to using the method in SciPy directly!
Example 2: fit of a decaying sine wave
In this example, we will explain some of the options of the algorithm.
We start off by generating some synthetic data with noise for a decaying sine wave, define an objective function, and create/initialize a Parameter set.
x = np.linspace(0, 15, 301)
np.random.seed(7)
noise = np.random.normal(size=x.size, scale=0.2)
data = (5. * np.sin(2*x - 0.1) * np.exp(-x*x*0.025) + noise)
plt.plot(x, data, 'o')
plt.show()
def fcn2min(params, x, data):
"""Model decaying sine wave, subtract data."""
amp = params['amp']
shift = params['shift']
omega = params['omega']
decay = params['decay']
model = amp * np.sin(x*omega + shift) * np.exp(-x*x*decay)
return model - data
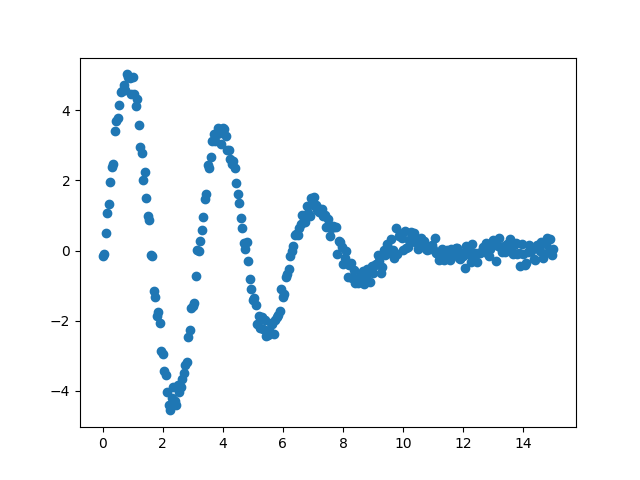
In contrast to the implementation in SciPy (as shown in the first example),
varying parameters do not need to have finite bounds in lmfit. However, if a
parameter does not have finite bounds, then it does need a brute_step
attribute specified:
Our initial parameter set is now defined as shown below and this will determine how the grid is set-up.
params.pretty_print()
Name Value Min Max Stderr Vary Expr Brute_Step
amp 7 2.5 inf None True None 0.25
decay 0.05 -inf inf None True None 0.005
omega 3 -inf 5 None True None 0.25
shift 0 -1.571 1.571 None True None None
First, we initialize a Minimizer and perform the grid search:
[[Fit Statistics]]
# fitting method = brute
# function evals = 375000
# data points = 301
# variables = 4
chi-square = 11.9353671
reduced chi-square = 0.04018642
Akaike info crit = -963.508878
Bayesian info crit = -948.680437
## Warning: uncertainties could not be estimated:
[[Variables]]
amp: 5.00000000 (init = 7)
decay: 0.02500000 (init = 0.05)
shift: -0.13089969 (init = 0)
omega: 2.00000000 (init = 3)
We used two new parameters here: Ns and keep. The parameter Ns
determines the 'number of grid points along the axes' similarly to its usage
in SciPy. Together with brute_step, min and max for a Parameter
it will dictate how the grid is set-up:
(1) finite bounds are specified (“SciPy implementation”): uses
brute_step if present (in the example above) or uses Ns to generate
the grid. The latter scenario that interpolates Ns points from min
to max (inclusive), is here shown for the parameter shift:
par_name = 'shift'
indx_shift = result_brute.var_names.index(par_name)
grid_shift = np.unique(result_brute.brute_grid[indx_shift].ravel())
print(f"parameter = {par_name}\nnumber of steps = {len(grid_shift)}\ngrid = {grid_shift}")
parameter = shift
number of steps = 25
grid = [-1.57079633 -1.43989663 -1.30899694 -1.17809725 -1.04719755 -0.91629786
-0.78539816 -0.65449847 -0.52359878 -0.39269908 -0.26179939 -0.13089969
0. 0.13089969 0.26179939 0.39269908 0.52359878 0.65449847
0.78539816 0.91629786 1.04719755 1.17809725 1.30899694 1.43989663
1.57079633]
If finite bounds are not set for a certain parameter then the user must
specify brute_step - three more scenarios are considered here:
(2) lower bound (min) and brute_step are specified:
range = (min, min + Ns * brute_step, brute_step)
par_name = 'amp'
indx_shift = result_brute.var_names.index(par_name)
grid_shift = np.unique(result_brute.brute_grid[indx_shift].ravel())
print(f"parameter = {par_name}\nnumber of steps = {len(grid_shift)}\ngrid = {grid_shift}")
parameter = amp
number of steps = 25
grid = [2.5 2.75 3. 3.25 3.5 3.75 4. 4.25 4.5 4.75 5. 5.25 5.5 5.75
6. 6.25 6.5 6.75 7. 7.25 7.5 7.75 8. 8.25 8.5 ]
(3) upper bound (max) and brute_step are specified:
range = (max - Ns * brute_step, max, brute_step)
par_name = 'omega'
indx_shift = result_brute.var_names.index(par_name)
grid_shift = np.unique(result_brute.brute_grid[indx_shift].ravel())
print(f"parameter = {par_name}\nnumber of steps = {len(grid_shift)}\ngrid = {grid_shift}")
parameter = omega
number of steps = 25
grid = [-1.25 -1. -0.75 -0.5 -0.25 0. 0.25 0.5 0.75 1. 1.25 1.5
1.75 2. 2.25 2.5 2.75 3. 3.25 3.5 3.75 4. 4.25 4.5
4.75]
(4) numerical value (value) and brute_step are specified:
range = (value - (Ns//2) * brute_step, value + (Ns//2) * brute_step, brute_step)
par_name = 'decay'
indx_shift = result_brute.var_names.index(par_name)
grid_shift = np.unique(result_brute.brute_grid[indx_shift].ravel())
print(f"parameter = {par_name}\nnumber of steps = {len(grid_shift)}\ngrid = {grid_shift}")
parameter = decay
number of steps = 24
grid = [-1.00000000e-02 -5.00000000e-03 5.20417043e-18 5.00000000e-03
1.00000000e-02 1.50000000e-02 2.00000000e-02 2.50000000e-02
3.00000000e-02 3.50000000e-02 4.00000000e-02 4.50000000e-02
5.00000000e-02 5.50000000e-02 6.00000000e-02 6.50000000e-02
7.00000000e-02 7.50000000e-02 8.00000000e-02 8.50000000e-02
9.00000000e-02 9.50000000e-02 1.00000000e-01 1.05000000e-01]
The MinimizerResult contains all the usual best-fit parameters and
fitting statistics. For example, the optimal solution from the grid search
is given below together with a plot:
print(fit_report(result_brute))
[[Fit Statistics]]
# fitting method = brute
# function evals = 375000
# data points = 301
# variables = 4
chi-square = 11.9353671
reduced chi-square = 0.04018642
Akaike info crit = -963.508878
Bayesian info crit = -948.680437
## Warning: uncertainties could not be estimated:
[[Variables]]
amp: 5.00000000 (init = 7)
decay: 0.02500000 (init = 0.05)
shift: -0.13089969 (init = 0)
omega: 2.00000000 (init = 3)
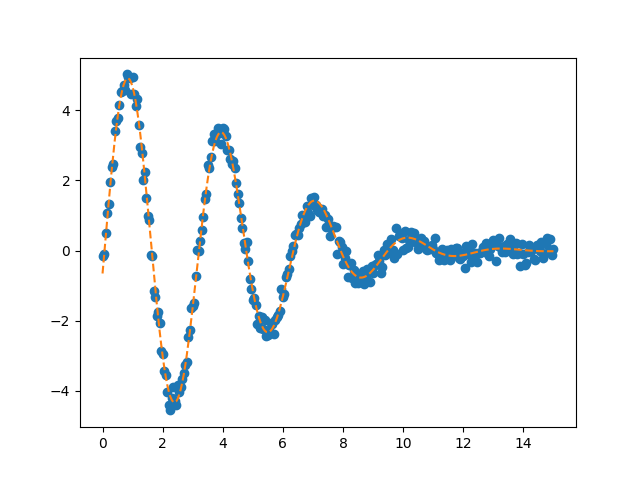
We can see that this fit is already very good, which is what we should expect
since our brute force grid is sampled rather finely and encompasses the
“correct” values.
In a more realistic, complicated example the brute method will be used
to get reasonable values for the parameters and perform another minimization
(e.g., using leastsq) using those as starting values. That is where the
keep parameter comes into play: it determines the “number of best
candidates from the brute force method that are stored in the candidates
attribute”. In the example above we store the best-ranking 25 solutions (the
default value is 50 and storing all the grid points can be accomplished
by choosing all). The candidates attribute contains the parameters
and chisqr from the brute force method as a namedtuple,
(‘Candidate’, [‘params’, ‘score’]), sorted on the (lowest) chisqr
value. To access the values for a particular candidate one can use
result.candidate[#].params or result.candidate[#].score, where a
lower # represents a better candidate. The show_candidates(#) uses the
pretty_print() method to show a specific candidate-# or all candidates
when no number is specified.
The optimal fit is, as usual, stored in the MinimizerResult.params
attribute and is, therefore, identical to result_brute.show_candidates(1).
result_brute.show_candidates(1)
Candidate #1, chisqr = 11.935
Name Value Min Max Stderr Vary Expr Brute_Step
amp 5 2.5 inf None True None 0.25
decay 0.025 -inf inf None True None 0.005
omega 2 -inf 5 None True None 0.25
shift -0.1309 -1.571 1.571 None True None None
In this case, the next-best scoring candidate has already a chisqr that
increased quite a bit:
result_brute.show_candidates(2)
Candidate #2, chisqr = 13.994
Name Value Min Max Stderr Vary Expr Brute_Step
amp 4.75 2.5 inf None True None 0.25
decay 0.025 -inf inf None True None 0.005
omega 2 -inf 5 None True None 0.25
shift -0.1309 -1.571 1.571 None True None None
and is, therefore, probably not so likely… However, as said above, in most
cases you’ll want to do another minimization using the solutions from the
brute method as starting values. That can be easily accomplished as
shown in the code below, where we now perform a leastsq minimization
starting from the top-25 solutions and accept the solution if the chisqr
is lower than the previously ‘optimal’ solution:
best_result = copy.deepcopy(result_brute)
for candidate in result_brute.candidates:
trial = fitter.minimize(method='leastsq', params=candidate.params)
if trial.chisqr < best_result.chisqr:
best_result = trial
From the leastsq minimization we obtain the following parameters for the
most optimal result:
print(fit_report(best_result))
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 21
# data points = 301
# variables = 4
chi-square = 10.8653514
reduced chi-square = 0.03658367
Akaike info crit = -991.780924
Bayesian info crit = -976.952483
[[Variables]]
amp: 5.00323085 +/- 0.03805940 (0.76%) (init = 5)
decay: 0.02563850 +/- 4.4572e-04 (1.74%) (init = 0.03)
shift: -0.09162987 +/- 0.00978382 (10.68%) (init = 0)
omega: 1.99611629 +/- 0.00316225 (0.16%) (init = 2)
[[Correlations]] (unreported correlations are < 0.100)
C(shift, omega) = -0.7855
C(amp, decay) = +0.5838
C(amp, shift) = -0.1208
As expected the parameters have not changed significantly as they were already very close to the “real” values, which can also be appreciated from the plots below.
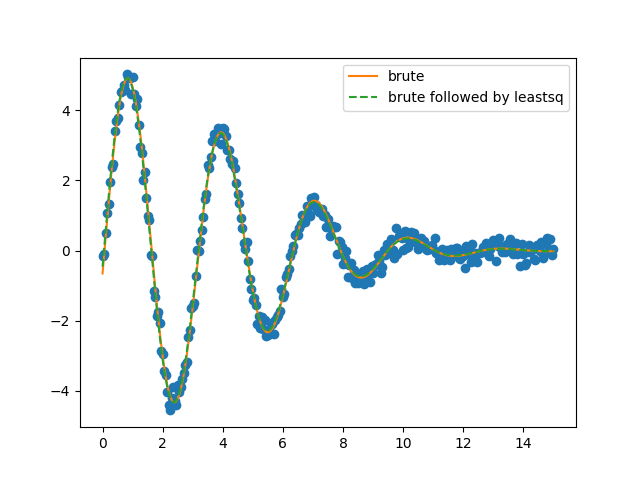
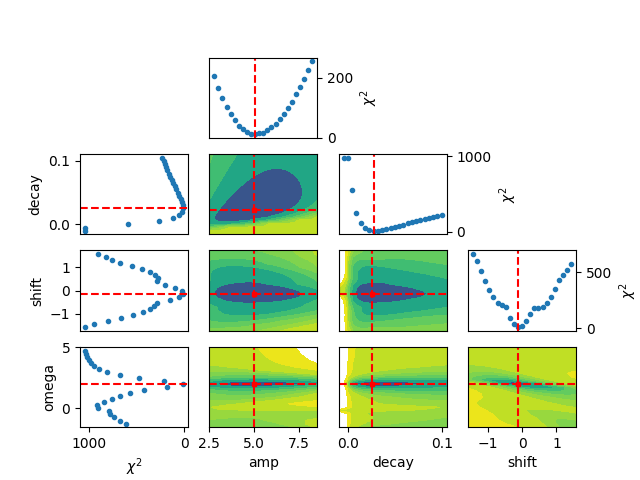
Finally, the results from the brute force grid-search can be visualized
using the rather lengthy Python function below (which might get incorporated
in lmfit at some point).
def plot_results_brute(result, best_vals=True, varlabels=None,
output=None):
"""Visualize the result of the brute force grid search.
The output file will display the chi-square value per parameter and contour
plots for all combination of two parameters.
Inspired by the `corner` package (https://github.com/dfm/corner.py).
Parameters
----------
result : :class:`~lmfit.minimizer.MinimizerResult`
Contains the results from the :meth:`brute` method.
best_vals : bool, optional
Whether to show the best values from the grid search (default is True).
varlabels : list, optional
If None (default), use `result.var_names` as axis labels, otherwise
use the names specified in `varlabels`.
output : str, optional
Name of the output PDF file (default is 'None')
"""
npars = len(result.var_names)
_fig, axes = plt.subplots(npars, npars)
if not varlabels:
varlabels = result.var_names
if best_vals and isinstance(best_vals, bool):
best_vals = result.params
for i, par1 in enumerate(result.var_names):
for j, par2 in enumerate(result.var_names):
# parameter vs chi2 in case of only one parameter
if npars == 1:
axes.plot(result.brute_grid, result.brute_Jout, 'o', ms=3)
axes.set_ylabel(r'$\chi^{2}$')
axes.set_xlabel(varlabels[i])
if best_vals:
axes.axvline(best_vals[par1].value, ls='dashed', color='r')
# parameter vs chi2 profile on top
elif i == j and j < npars-1:
if i == 0:
axes[0, 0].axis('off')
ax = axes[i, j+1]
red_axis = tuple(a for a in range(npars) if a != i)
ax.plot(np.unique(result.brute_grid[i]),
np.minimum.reduce(result.brute_Jout, axis=red_axis),
'o', ms=3)
ax.set_ylabel(r'$\chi^{2}$')
ax.yaxis.set_label_position("right")
ax.yaxis.set_ticks_position('right')
ax.set_xticks([])
if best_vals:
ax.axvline(best_vals[par1].value, ls='dashed', color='r')
# parameter vs chi2 profile on the left
elif j == 0 and i > 0:
ax = axes[i, j]
red_axis = tuple(a for a in range(npars) if a != i)
ax.plot(np.minimum.reduce(result.brute_Jout, axis=red_axis),
np.unique(result.brute_grid[i]), 'o', ms=3)
ax.invert_xaxis()
ax.set_ylabel(varlabels[i])
if i != npars-1:
ax.set_xticks([])
else:
ax.set_xlabel(r'$\chi^{2}$')
if best_vals:
ax.axhline(best_vals[par1].value, ls='dashed', color='r')
# contour plots for all combinations of two parameters
elif j > i:
ax = axes[j, i+1]
red_axis = tuple(a for a in range(npars) if a not in (i, j))
X, Y = np.meshgrid(np.unique(result.brute_grid[i]),
np.unique(result.brute_grid[j]))
lvls1 = np.linspace(result.brute_Jout.min(),
np.median(result.brute_Jout)/2.0, 7, dtype='int')
lvls2 = np.linspace(np.median(result.brute_Jout)/2.0,
np.median(result.brute_Jout), 3, dtype='int')
lvls = np.unique(np.concatenate((lvls1, lvls2)))
ax.contourf(X.T, Y.T, np.minimum.reduce(result.brute_Jout, axis=red_axis),
lvls, norm=LogNorm())
ax.set_yticks([])
if best_vals:
ax.axvline(best_vals[par1].value, ls='dashed', color='r')
ax.axhline(best_vals[par2].value, ls='dashed', color='r')
ax.plot(best_vals[par1].value, best_vals[par2].value, 'rs', ms=3)
if j != npars-1:
ax.set_xticks([])
else:
ax.set_xlabel(varlabels[i])
if j - i >= 2:
axes[i, j].axis('off')
if output is not None:
plt.savefig(output)
and finally, to generated the figure:
plot_results_brute(result_brute, best_vals=True, varlabels=None)
plt.show()
Total running time of the script: (0 minutes 19.744 seconds)
