Note
Go to the end to download the full example code.
Fit Using differential_evolution Algorithm¶
This example compares the leastsq and differential_evolution algorithms
on a fairly simple problem.
Generate synthetic data and set-up Parameters with initial values/boundaries:
decay = 5
offset = 1.0
amp = 2.0
omega = 4.0
np.random.seed(2)
x = np.linspace(0, 10, 101)
y = offset + amp*np.sin(omega*x) * np.exp(-x/decay)
yn = y + np.random.normal(size=y.size, scale=0.450)
params = lmfit.Parameters()
params.add('offset', 2.0, min=0, max=10.0)
params.add('omega', 3.3, min=0, max=10.0)
params.add('amp', 2.5, min=0, max=10.0)
params.add('decay', 1.0, min=0, max=10.0)
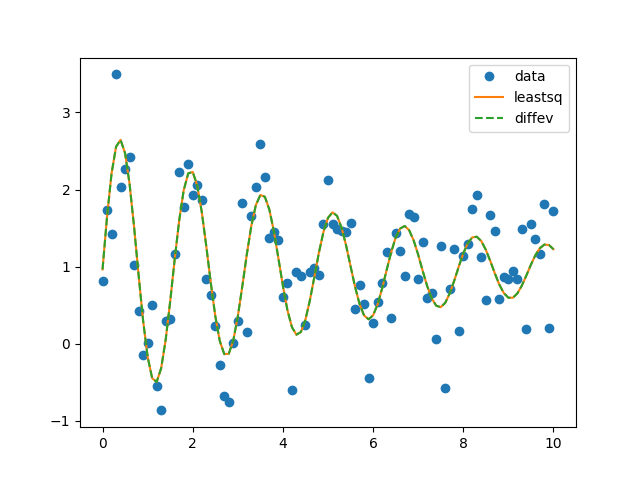
Perform the fits and show fitting results and plot:
# Fit using leastsq:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 65
# data points = 101
# variables = 4
chi-square = 21.7961792
reduced chi-square = 0.22470288
Akaike info crit = -146.871969
Bayesian info crit = -136.411487
[[Variables]]
offset: 0.96333089 +/- 0.04735890 (4.92%) (init = 2)
omega: 3.98700839 +/- 0.02079709 (0.52%) (init = 3.3)
amp: 1.80253587 +/- 0.19401928 (10.76%) (init = 2.5)
decay: 5.76279753 +/- 1.04073348 (18.06%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amp, decay) = -0.7550
# Fit using differential_evolution:
[[Fit Statistics]]
# fitting method = differential_evolution
# function evals = 1720
# data points = 101
# variables = 4
chi-square = 21.7961792
reduced chi-square = 0.22470288
Akaike info crit = -146.871969
Bayesian info crit = -136.411487
[[Variables]]
offset: 0.96333168 +/- 0.04735902 (4.92%) (init = 2)
omega: 3.98700858 +/- 0.02121811 (0.53%) (init = 3.3)
amp: 1.80253519 +/- 0.19022457 (10.55%) (init = 2.5)
decay: 5.76281240 +/- 1.00451705 (17.43%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amp, decay) = -0.7434
plt.plot(x, yn, 'o', label='data')
plt.plot(x, yn+o1.residual, '-', label='leastsq')
plt.plot(x, yn+o2.residual, '--', label='diffev')
plt.legend()
Total running time of the script: (0 minutes 0.474 seconds)
